Structural Characterization of Mycobacterium tuberculosis Encapsulin in Complex with Dye-Decolorizing Peroxide.
Cuthbert, B.J., Chen, X., Burley, K., Batot, G., Contreras, H., Dixon, S., Goulding, C.W.(2024) Microorganisms 12
- PubMed: 39770668
- DOI: https://doi.org/10.3390/microorganisms12122465
- Primary Citation of Related Structures:
9BKX - PubMed Abstract:
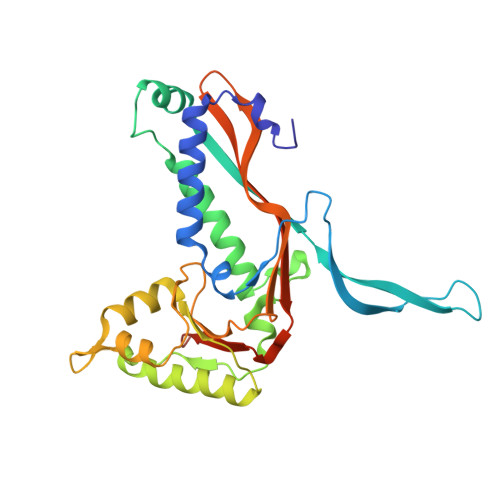
Mycobacterium tuberculosis (Mtb) is the causative agent of tuberculosis, the world's deadliest infectious disease. Mtb uses a variety of mechanisms to evade the human host's defenses and survive intracellularly. Mtb's oxidative stress response enables Mtb to survive within activated macrophages, an environment with reactive oxygen species and low pH. Dye-decolorizing peroxidase (DyP), an enzyme involved in Mtb's oxidative stress response, is encapsulated in a nanocompartment, encapsulin (Enc), and is important for Mtb's survival in macrophages. Encs are homologs of viral capsids and encapsulate cargo proteins of diverse function, including those involved in iron storage and stress responses. DyP contains a targeting peptide (TP) at its C-terminus that recognizes and binds to the interior of the Enc nanocompartment. Here, we present the crystal structure of the Mtb-Enc•DyP complex and compare it to cryogenic-electron microscopy (cryo-EM) Mtb-Enc structures. Investigation into the canonical pores formed at symmetrical interfaces reveals that the five-fold pore for the Mtb-Enc crystal structure is strikingly different from that observed in cryo-EM structures. We also observe DyP-TP electron density within the Mtb-Enc shell. Finally, investigation into crystallographic small-molecule binding sites gives insight into potential novel avenues by which substrates could enter Mtb-Enc to react with Mtb-DyP.
- Department of Molecular Biology and Biochemistry, University of California Irvine, Irvine, CA 92697, USA.
Organizational Affiliation: