PROTAC-mediated activation, rather than degradation, of a nuclear receptor reveals complex ligand-receptor interaction network.
Huber, A.D., Lin, W., Poudel, S., Miller, D.J., Chen, T.(2024) Structure 32: 2352-2363.e8
- PubMed: 39389062
- DOI: https://doi.org/10.1016/j.str.2024.09.016
- Primary Citation of Related Structures:
9BEQ - PubMed Abstract:
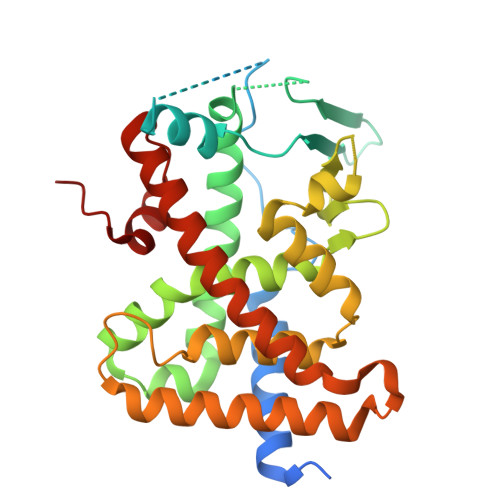
Proteolysis-targeting chimeras (PROTACs) are heterobifunctional molecules containing a ligand for a protein of interest linked to an E3 ubiquitin ligase ligand that induce protein degradation through E3 recruitment to the target protein. Small changes in PROTAC linkers can have drastic consequences, including loss of degradation activity, but the structural mechanisms governing such changes are unclear. To study this phenomenon, we screened PROTACs of diverse targeting modalities and identified dTAG-13 as an activator of the xenobiotic-sensing pregnane X receptor (PXR), which promiscuously binds various ligands. Characterization of dTAG-13 analogs and precursors revealed interplay between the PXR-binding moiety, linker, and E3 ligand that altered PXR activity without inducing degradation. A crystal structure of PXR ligand binding domain bound to a precursor ligand showed ligand-induced binding pocket distortions and a linker-punctured tunnel to the protein exterior at a region incompatible with E3 complex formation, highlighting the effects of linker environment on PROTAC activity.
- Department of Chemical Biology and Therapeutics, St. Jude Children's Research Hospital, 262 Danny Thomas Place, MS 1000, Memphis, TN 38105-3678, USA.
Organizational Affiliation: