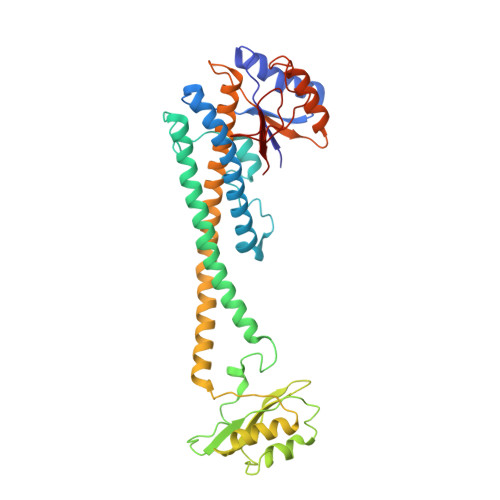
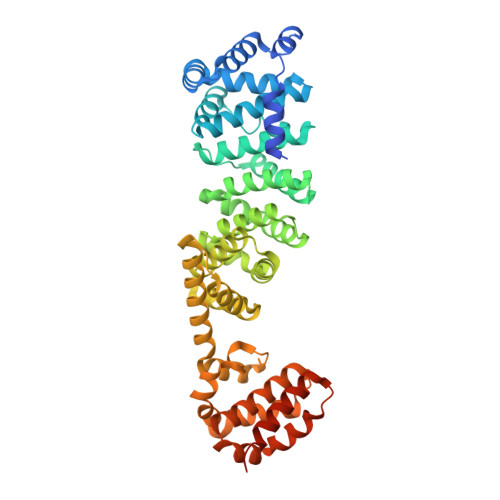
High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Coupland, C.E., Karimi, R., Bueler, S.A., Liang, Y., Courbon, G.M., Di Trani, J.M., Wong, C.J., Saghian, R., Youn, J.Y., Wang, L.Y., Rubinstein, J.L.(2024) Science 385: 168-174
- PubMed: 38900912
- DOI: https://doi.org/10.1126/science.adp5577
- Primary Citation of Related Structures:
9B8O, 9B8P, 9B8Q, 9BRB, 9BRC, 9BRD - PubMed Abstract:
Intercellular communication in the nervous system occurs through the release of neurotransmitters into the synaptic cleft between neurons. In the presynaptic neuron, the proton pumping vesicular- or vacuolar-type ATPase (V-ATPase) powers neurotransmitter loading into synaptic vesicles (SVs), with the V 1 complex dissociating from the membrane region of the enzyme before exocytosis. We isolated SVs from rat brain using SidK, a V-ATPase-binding bacterial effector protein. Single particle electron cryomicroscopy allowed high-resolution structure determination of V-ATPase within the native SV membrane. In the structure, regularly spaced cholesterol molecules decorate the enzyme's rotor and the abundant SV protein synaptophysin binds the complex stoichiometrically. ATP hydrolysis during vesicle loading results in loss of V 1 from the SV membrane, suggesting that loading is sufficient to induce dissociation of the enzyme.
- Molecular Medicine Program, The Hospital for Sick Children, Toronto, Canada M5G 1X8.
Organizational Affiliation: