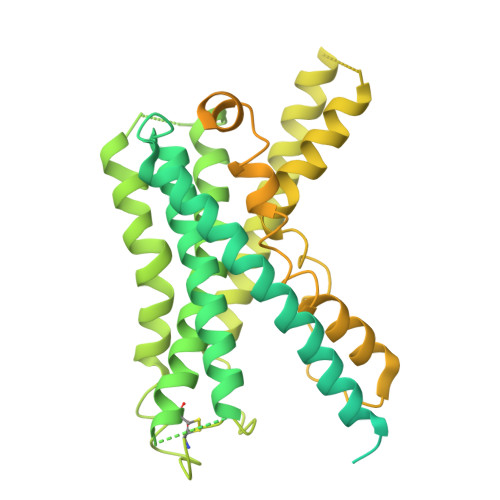
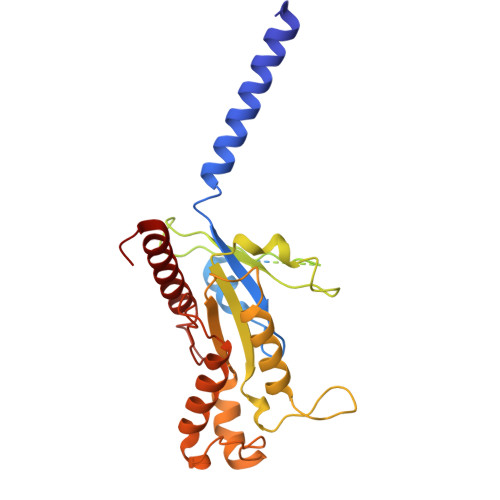
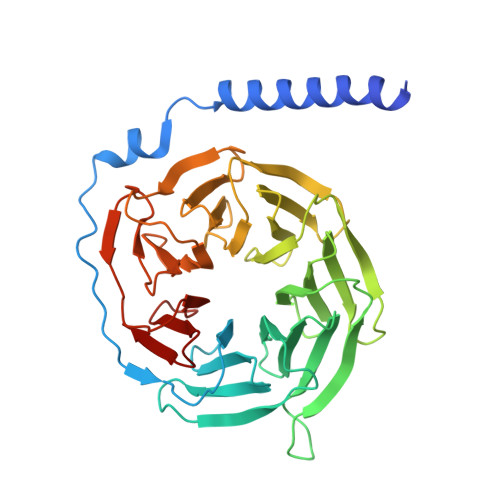
Allosteric mechanism in the distinctive coupling of G q and G s to the parathyroid hormone type 1 receptor.
Zhang, X., Lee, J.Y., Pacheco, J., Sutkeviciute, I., Anitha, A.K., Liu, H., Singh, S., Ventura, C., Savransky, S., Khatri, A., Zhang, C., Bahar, I., Vilardaga, J.P.(2025) Proc Natl Acad Sci U S A 122: e2426178122-e2426178122
- PubMed: 40138341
- DOI: https://doi.org/10.1073/pnas.2426178122
- Primary Citation of Related Structures:
9B5Y - PubMed Abstract:
The mechanism determining the preferential stimulation of one heterotrimeric G protein signaling pathway over another by a ligand remains undetermined. By reporting the cryogenic electron microscopy (cryo-EM) structure of the parathyroid hormone (PTH) type 1 receptor (PTH1R) complexed with Gq and comparing its allosteric dynamics with that of PTH1R in complex with G s , we uncover a mechanism underlying such preferences. We show that an allosteric coupling between the ligand PTH and the C-terminal helix α5 of the Gα subunit controls the stability of the PTH1R complex with the specific G protein, G s or G q . Single-cell-level experiments further validate the G protein-selective effects of the PTH binding pose by demonstrating the differential, G protein-dependent residence times and affinity of this ligand at the PTH1R binding site. The findings deepen our understanding of the selective coupling of PTH1R to G s or G q and how it relates to the stability and kinetics of ligand binding. They explain the observed variability in the ligand-binding affinity of a GPCR when coupled to different G proteins.
- Department of Pharmacology and Chemical Biology, School of Medicine, University of Pittsburgh, Pittsburgh, PA 15261.
Organizational Affiliation: