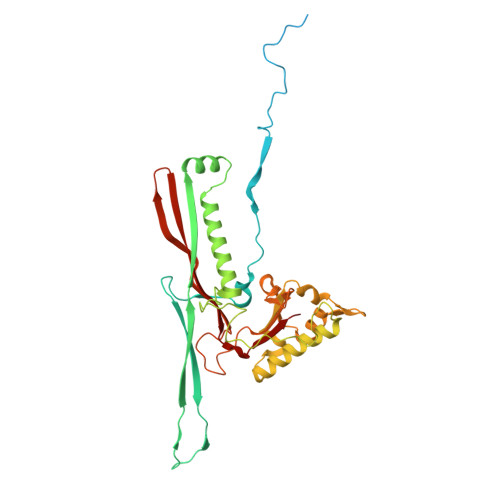
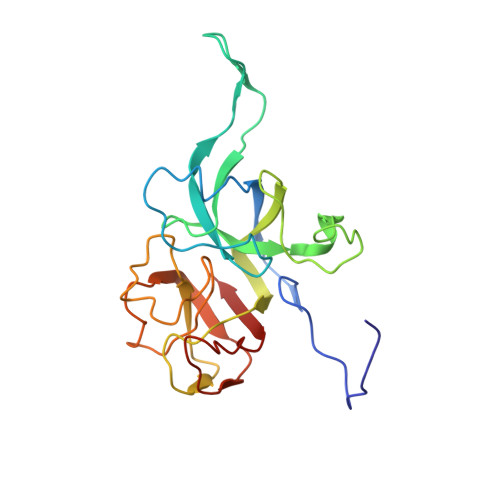
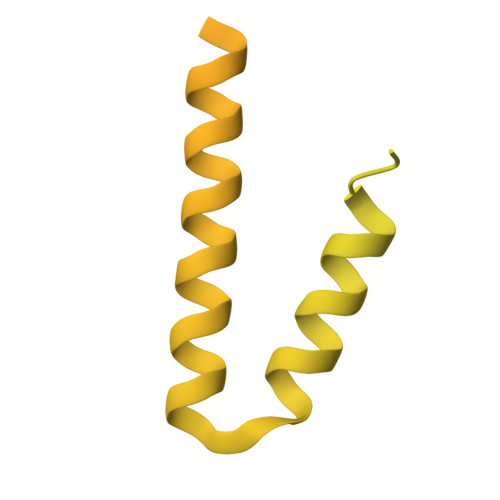
Cryo-EM analysis of Pseudomonas phage Pa193 structural components.
Iglesias, S.M., Hou, C.D., Reid, J., Schauer, E., Geier, R., Soriaga, A., Sim, L., Gao, L., Whitelegge, J., Kyme, P., Birx, D., Lemire, S., Cingolani, G.(2024) Commun Biol 7: 1275-1275
- PubMed: 39370451
- DOI: https://doi.org/10.1038/s42003-024-06985-x
- Primary Citation of Related Structures:
9B40, 9B41, 9B42, 9B45 - PubMed Abstract:
The World Health Organization has designated Pseudomonas aeruginosa as a critical pathogen for the development of new antimicrobials. Bacterial viruses, or bacteriophages, have been used in various clinical settings, commonly called phage therapy, to address this growing public health crisis. Here, we describe a high-resolution structural atlas of a therapeutic, contractile-tailed Pseudomonas phage, Pa193. We used bioinformatics, proteomics, and cryogenic electron microscopy single particle analysis to identify, annotate, and build atomic models for 21 distinct structural polypeptide chains forming the icosahedral capsid, neck, contractile tail, and baseplate. We identified a putative scaffolding protein stabilizing the interior of the capsid 5-fold vertex. We also visualized a large portion of Pa193 ~ 500 Å long tail fibers and resolved the interface between the baseplate and tail fibers. The work presented here provides a framework to support a better understanding of phages as biomedicines for phage therapy and inform engineering opportunities.
- Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, USA.
Organizational Affiliation: