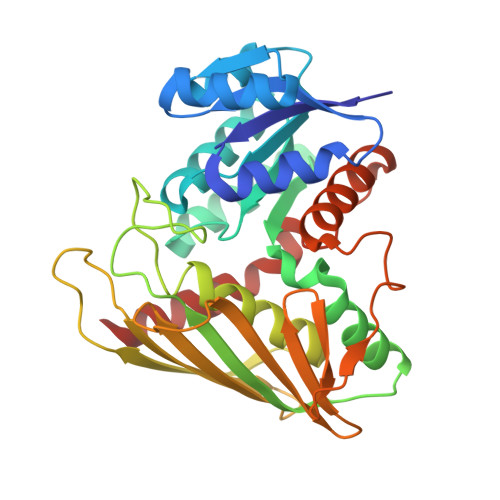
Characterization of lignin-degrading enzyme PmdC, which catalyzes a key step in the synthesis of polymer precursor 2-pyrone-4,6-dicarboxylic acid.
Rodrigues, A.V., Moriarty, N.W., Kakumanu, R., DeGiovanni, A., Pereira, J.H., Gin, J.W., Chen, Y., Baidoo, E.E.K., Petzold, C.J., Adams, P.D.(2024) J Biological Chem 300: 107736-107736
- PubMed: 39222681
- DOI: https://doi.org/10.1016/j.jbc.2024.107736
- Primary Citation of Related Structures:
9AZO - PubMed Abstract:
Pyrone-2,4-dicarboxylic acid (PDC) is a valuable polymer precursor that can be derived from the microbial degradation of lignin. The key enzyme in the microbial production of PDC is CHMS dehydrogenase, which acts on the substrate 4-carboxy-2-hydroxymuconate-6-semialdehyde (CHMS). We present the crystal structure of CHMS dehydrogenase (PmdC from Comamonas testosteroni) bound to the cofactor NADP, shedding light on its three-dimensional architecture, and revealing residues responsible for binding NADP. Using a combination of structural homology, molecular docking, and quantum chemistry calculations we have predicted the binding site of CHMS. Key histidine residues in a conserved sequence are identified as crucial for binding the hydroxyl group of CHMS and facilitating dehydrogenation with NADP. Mutating these histidine residues results in a loss of enzyme activity, leading to a proposed model for the enzyme's mechanism. These findings are expected to help guide efforts in protein and metabolic engineering to enhance PDC yields in biological routes to polymer feedstock synthesis.
- Joint BioEnergy Institute, Emeryville, California, 94608, United States; Molecular Biophysics and Integrated Bioimaging, Lawrence Berkeley National Laboratory, Berkeley California 94720, United States. Electronic address: avrodrigues@lbl.gov.
Organizational Affiliation: