Structural basis of error-prone DNA synthesis by DNA polymerase theta.
Li, C., Maksoud, L.M., Gao, Y.(2025) Nat Commun 16: 2063-2063
- PubMed: 40021647
- DOI: https://doi.org/10.1038/s41467-025-57269-9
- Primary Citation of Related Structures:
9AU5, 9AU8, 9AU9 - PubMed Abstract:
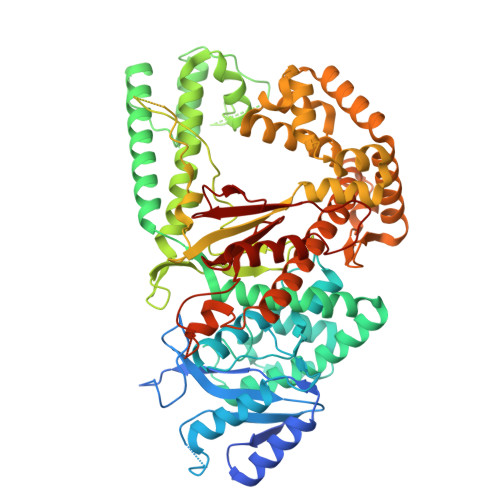
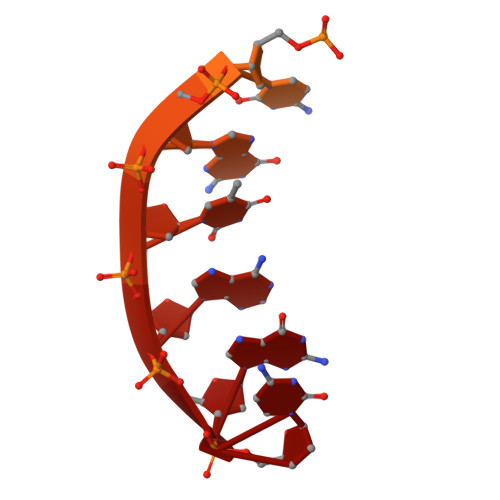
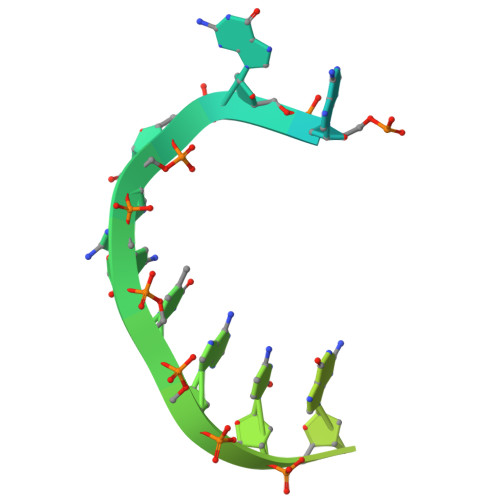
DNA polymerase θ (Pol θ) is an A-family DNA polymerase specialized in DNA double-strand breaks repair and translesion synthesis. Distinct from its high-fidelity homologs in DNA replication, Pol θ catalyzes template-dependent DNA synthesis with an inherent propensity for error incorporation. However, the structural basis of Pol θ's low-fidelity DNA synthesis is not clear. Here, we present cryo-electron microscopy structures detailing the polymerase domain of human Pol θ in complex with a cognate C:G base pair (bp), a mismatched T:G bp, or a mismatched T:T bp. Our structures illustrate that Pol θ snugly accommodates the mismatched nascent base pairs within its active site with the finger domain well-closed, consistent with our in-solution fluorescence measurement but in contrast to its high-fidelity homologs. In addition, structural examination and mutagenesis study show that unique residues surrounding the active site contribute to the stabilization of the mismatched nascent base pair. Furthermore, Pol θ can efficiently extend from the misincorporated T:G or T:T mismatches, yet with a preference for template or primer looping-out, resulting in insertions and deletions. Collectively, our results elucidate how an A-family polymerase is adapted for error-prone DNA synthesis.
- Department of Biosciences, Rice University, 6500 Main St., Houston, 77005, TX, USA.
Organizational Affiliation: