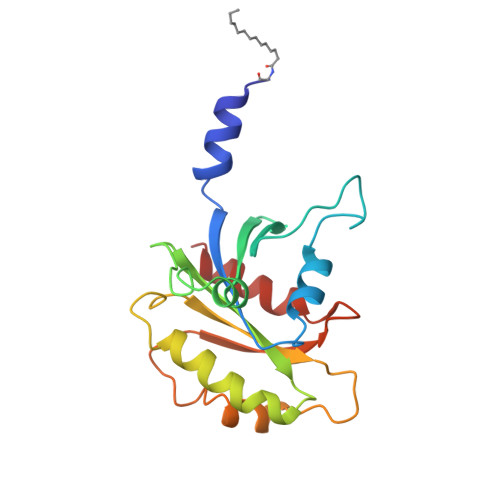
Myr-Arf1 conformational flexibility at the membrane surface sheds light on the interactions with ArfGAP ASAP1
Zhang, Y., Soubias, O., Pant, S., Heinrich, F., Vogel, A., Li, J., Li, Y., Clifton, L.A., Daum, S., Bacia, K., Huster, D., Randazzo, P.A., Losche, M., Tajkhorshid, E., Byrd, R.A.(2023) Nat Commun
- PubMed: 37989735
- DOI: https://doi.org/10.1038/s41467-023-43008-5
- Primary Citation of Related Structures:
9A8S - PubMed Abstract:
ADP-ribosylation factor 1 (Arf1) interacts with multiple cellular partners and membranes to regulate intracellular traffic, organelle structure and actin dynamics. Defining the dynamic conformational landscape of Arf1 in its active form, when bound to the membrane, is of high functional relevance and key to understanding how Arf1 can alter diverse cellular processes. Through concerted application of nuclear magnetic resonance (NMR), neutron reflectometry (NR) and molecular dynamics (MD) simulations, we show that, while Arf1 is anchored to the membrane through its N-terminal myristoylated amphipathic helix, the G domain explores a large conformational space, existing in a dynamic equilibrium between membrane-associated and membrane-distal conformations. These configurational dynamics expose different interfaces for interaction with effectors. Interaction with the Pleckstrin homology domain of ASAP1, an Arf-GTPase activating protein (ArfGAP), restricts motions of the G domain to lock it in what seems to be a conformation exposing functionally relevant regions.
- Center for Structural Biology, Center for Cancer Research, National Cancer Institute, Frederick, MD, 21702-1201, USA.
Organizational Affiliation: