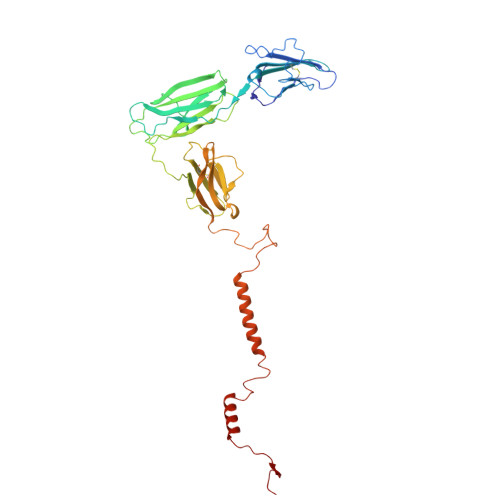
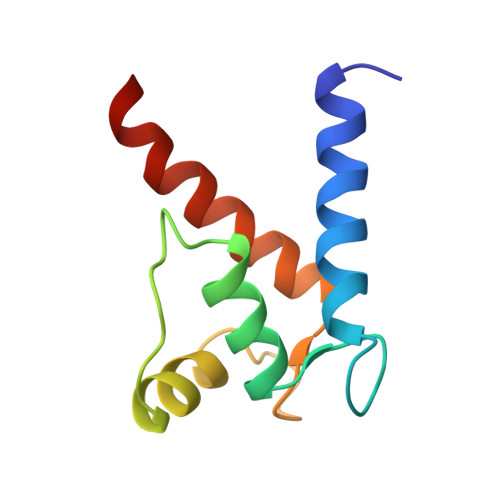
A model of full-length RAGE in complex with S100B
Moysa, A., Steczkiewicz, K., Niedzialek, D., Hammerschmid, D., Zhukova, L., Sobott, F., Dadlez, M.(2021) Structure 29: 989
- PubMed: 33887170
- DOI: https://doi.org/10.1016/j.str.2021.04.002
- Primary Citation of Related Structures:
9A18 - PubMed Abstract:
The receptor for advanced glycation end products (RAGE) is an immunoglobulin-type multiligand transmembrane protein expressed in numerous cell types, including the central nervous system cells. RAGE interaction with S100B, released during brain tissue damage, leads to RAGE upregulation and initialization of a spiral proinflammatory associated with different neural disorders. Here, we present the structural characterization of the hetero-oligomeric complex of the full-length RAGE with S100B, obtained by a combination of mass spectrometry-based methods and molecular modeling. We predict that RAGE functions as a tightly packed tetramer exposing a positively charged surface formed by V domains for S100B binding. Based on HDX results we demonstrate an allosteric coupling of the distal extracellular V domains and the transmembrane region, indicating a possible mechanism of signal transmission by RAGE across the membrane. Our model provides an insight into RAGE-ligand interactions, providing a basis for the rational design of the therapeutic modifiers of its activity.
- Institute of Biochemistry and Biophysics, PAS, Pawinskiego 5a, 02-109 Warsaw, Poland. Electronic address: a.alexandrmoysa@gmail.com.
Organizational Affiliation: