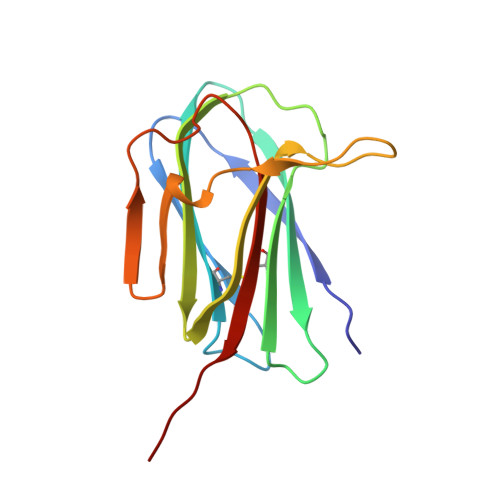
Solution structure of human myeloid-derived growth factor suggests a conserved function in the endoplasmic reticulum
Bortnov V, Tonelli M, Lee W, Lin Z, Annis DS, Demerdash ON, Bateman A, Mitchell JC, Ge Y, Markley JL, Mosher DF(2019) Nat Commun 10: 5612
- PubMed: 31819058
- DOI: https://doi.org/10.1038/s41467-019-13577-5
- Primary Citation of Related Structures:
6O6W, 9A00 - PubMed Abstract:
Human myeloid-derived growth factor (hMYDGF) is a 142-residue protein with a C-terminal endoplasmic reticulum (ER) retention sequence (ERS). Extracellular MYDGF mediates cardiac repair in mice after anoxic injury. Although homologs of hMYDGF are found in eukaryotes as distant as protozoans, its structure and function are unknown. Here we present the NMR solution structure of hMYDGF, which consists of a short α-helix and ten β-strands distributed in three β-sheets. Conserved residues map to the unstructured ERS, loops on the face opposite the ERS, and the surface of a cavity underneath the conserved loops. The only protein or portion of a protein known to have a similar fold is the base domain of VNN1. We suggest, in analogy to the tethering of the VNN1 nitrilase domain to the plasma membrane via its base domain, that MYDGF complexed to the KDEL receptor binds cargo via its conserved residues for transport to the ER.
- Department of Biomolecular Chemistry, University of Wisconsin-Madison, Madison, WI, 53706, USA.
Organizational Affiliation: