1.8 A resolution structure of the Photosystem I assembly intermediate lacking stromal subunits
Charras, Q., Gupta, S., Tichy, M., Al-Amoudi, A., Lukes, M., Konik, P., Sobotka, R., Novak, P., Komenda, J., Naschberger, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I P700 chlorophyll a apoprotein A1 | 751 | Synechocystis | Mutation(s): 0 EC: 1.97.1.12 | 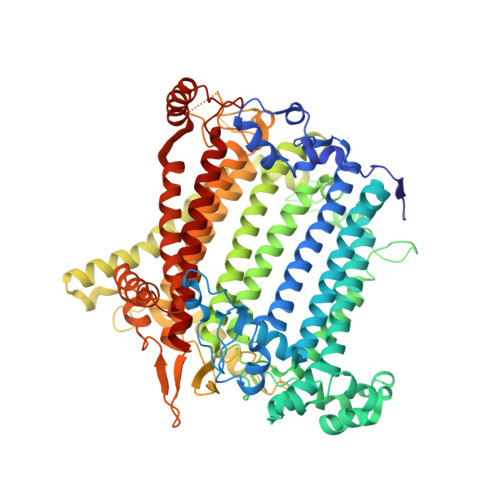 | |
UniProt | |||||
Find proteins for P29254 (Synechocystis sp. (strain ATCC 27184 / PCC 6803 / Kazusa)) Explore P29254 Go to UniProtKB: P29254 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P29254 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I P700 chlorophyll a apoprotein A2 | 731 | Synechocystis | Mutation(s): 0 EC: 1.97.1.12 | 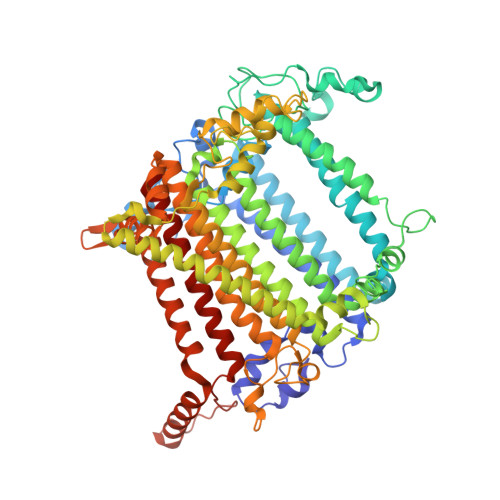 | |
UniProt | |||||
Find proteins for P29255 (Synechocystis sp. (strain ATCC 27184 / PCC 6803 / Kazusa)) Explore P29255 Go to UniProtKB: P29255 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P29255 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I reaction center subunit III | C [auth F] | 165 | Synechocystis | Mutation(s): 0 | 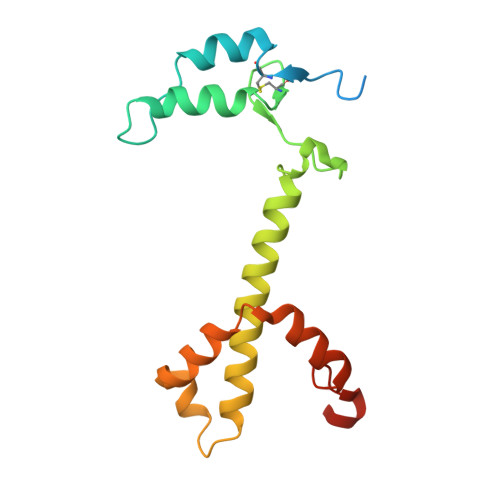 |
UniProt | |||||
Find proteins for P29256 (Synechocystis sp. (strain ATCC 27184 / PCC 6803 / Kazusa)) Explore P29256 Go to UniProtKB: P29256 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P29256 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I reaction center subunit VIII | D [auth I] | 40 | Synechocystis | Mutation(s): 0 | 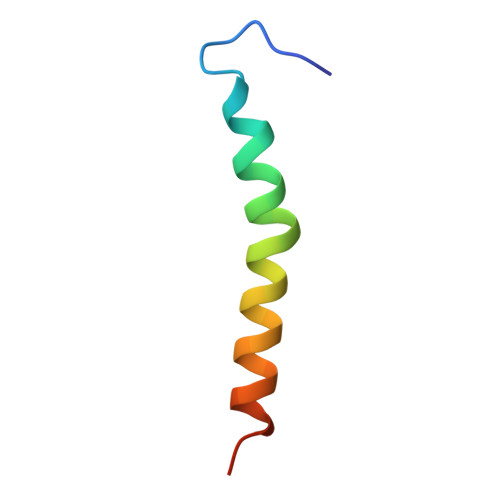 |
UniProt | |||||
Find proteins for Q55330 (Synechocystis sp. (strain ATCC 27184 / PCC 6803 / Kazusa)) Explore Q55330 Go to UniProtKB: Q55330 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q55330 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I reaction center subunit IX | E [auth J] | 40 | Synechocystis | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q55329 (Synechocystis sp. (strain ATCC 27184 / PCC 6803 / Kazusa)) Explore Q55329 Go to UniProtKB: Q55329 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q55329 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I reaction center subunit PsaK 1 | F [auth K] | 86 | Synechocystis | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P72712 (Synechocystis sp. (strain ATCC 27184 / PCC 6803 / Kazusa)) Explore P72712 Go to UniProtKB: P72712 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P72712 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I reaction center subunit XII | G [auth M] | 31 | Synechocystis | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P72986 (Synechocystis sp. (strain ATCC 27184 / PCC 6803 / Kazusa)) Explore P72986 Go to UniProtKB: P72986 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P72986 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 11 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CLA (Subject of Investigation/LOI) Query on CLA | AA [auth A] AB [auth A] AC [auth B] AD [auth B] BA [auth A] | CHLOROPHYLL A C55 H72 Mg N4 O5 ATNHDLDRLWWWCB-AENOIHSZSA-M |  | ||
| CL0 (Subject of Investigation/LOI) Query on CL0 | J [auth A] | CHLOROPHYLL A ISOMER C55 H72 Mg N4 O5 VIQFHHZSLDFWDU-DVXFRRMCSA-M |  | ||
| LMG (Subject of Investigation/LOI) Query on LMG | GB [auth A], SD [auth B] | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE C45 H86 O10 DCLTVZLYPPIIID-CVELTQQQSA-N |  | ||
| LHG (Subject of Investigation/LOI) Query on LHG | H [auth A], I [auth A], TD [auth B] | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE C38 H75 O10 P BIABMEZBCHDPBV-MPQUPPDSSA-N |  | ||
| 5X6 Query on 5X6 | UD [auth B] | Zeaxanthin C40 H56 O2 JKQXZKUSFCKOGQ-AYORJBSZSA-N |  | ||
| ECH Query on ECH | FB [auth A], IE [auth F], OD [auth B], TB [auth A], XE [auth M] | beta,beta-caroten-4-one C40 H54 O QXNWZXMBUKUYMD-QQGJMDNJSA-N |  | ||
| BCR Query on BCR | BB [auth A] CB [auth A] DB [auth A] EB [auth A] FE [auth F] | BETA-CAROTENE C40 H56 OENHQHLEOONYIE-JLTXGRSLSA-N |  | ||
| LMT (Subject of Investigation/LOI) Query on LMT | AE [auth B] BE [auth B] CE [auth B] DE [auth B] EE [auth F] | DODECYL-BETA-D-MALTOSIDE C24 H46 O11 NLEBIOOXCVAHBD-QKMCSOCLSA-N |  | ||
| PQN Query on PQN | M [auth A], YB [auth B] | PHYLLOQUINONE C31 H46 O2 MBWXNTAXLNYFJB-NKFFZRIASA-N |  | ||
| SF4 (Subject of Investigation/LOI) Query on SF4 | ZB [auth B] | IRON/SULFUR CLUSTER Fe4 S4 LJBDFODJNLIPKO-UHFFFAOYSA-N |  | ||
| CL Query on CL | QB [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Grant Agency of the Czech Republic | Czech Republic | 24-10227S |
| European Research Council (ERC) | European Union | 854126 |