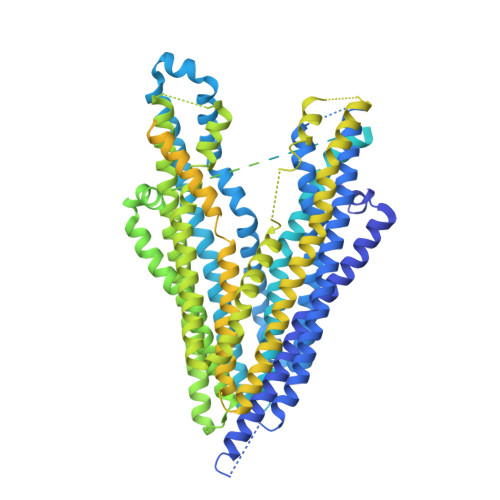
Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Hamaguchi-Suzuki, N., Adachi, N., Moriya, T., Yasuda, S., Kawasaki, M., Suzuki, K., Ogasawara, S., Anzai, N., Senda, T., Murata, T.(2024) Biochem Biophys Res Commun 709: 149855-149855
- PubMed: 38579618
- DOI: https://doi.org/10.1016/j.bbrc.2024.149855
- Primary Citation of Related Structures:
8Y6H, 8Y6I - PubMed Abstract:
P-glycoprotein (P-gp) is an ATP-binding cassette transporter known for its roles in expelling xenobiotic compounds from cells and contributing to cellular drug resistance through multidrug efflux. This mechanism is particularly problematic in cancer cells, where it diminishes the therapeutic efficacy of anticancer drugs. P-gp inhibitors, such as elacridar, have been developed to circumvent the decrease in drug efficacy due to P-gp efflux. An earlier study reported the cryo-EM structure of human P-gp-Fab (MRK-16) complex bound by two elacridar molecules, at a resolution of 3.6 Å. In this study, we have obtained a higher resolution (2.5 Å) structure of the P-gp- Fab (UIC2) complex bound by three elacridar molecules. This finding, which exposes a larger space for compound-binding sites than previously acknowledged, has significant implications for the development of more selective inhibitors and enhances our understanding of the compound recognition mechanism of P-gp.
- Department of Pharmacology, Chiba University Graduate School of Medicine, 1-8-1 Inohana, Chuo, Chiba, 260-8670, Japan; Department of Chemistry, Graduate School of Science, Chiba University, 1-33 Yayoi-cho, Inage, Chiba, 263-8522, Japan.
Organizational Affiliation: