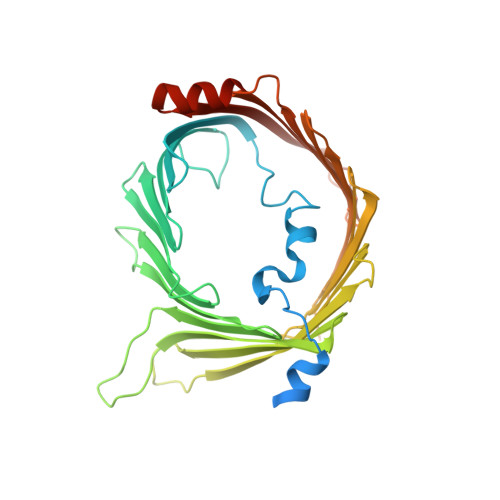
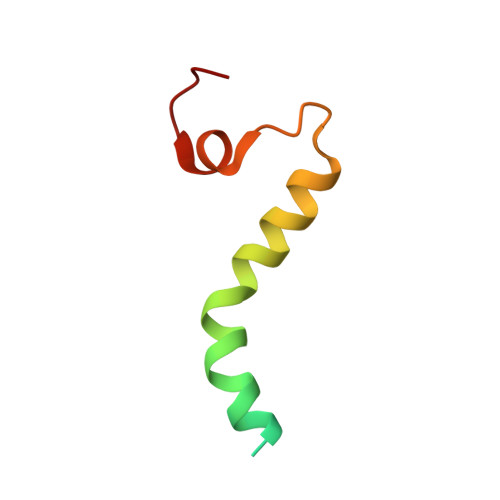
The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Wang, Q., Zhuang, J., Huang, R., Guan, Z., Yan, L., Hong, S., Zhang, L., Huang, C., Liu, Z., Yin, P.(2024) Cell Discov 10: 19-19
- PubMed: 38360717
- DOI: https://doi.org/10.1038/s41421-023-00643-y
- Primary Citation of Related Structures:
8W5J, 8W5K - National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei, China.
Organizational Affiliation: