Biochemical and structural characterization of Rab3GAP reveals insights into Rab18 nucleotide exchange activity.
Fairlie, G.M.J., Nguyen, K.M., Nam, S.E., Shaw, A.L., Parson, M.A.H., Shariati, H.R., Wang, X., Jenkins, M.L., Gong, M., Burke, J.E., Yip, C.K.(2025) Nat Commun 16: 479-479
- PubMed: 39779760
- DOI: https://doi.org/10.1038/s41467-025-55828-8
- Primary Citation of Related Structures:
8VYB - PubMed Abstract:
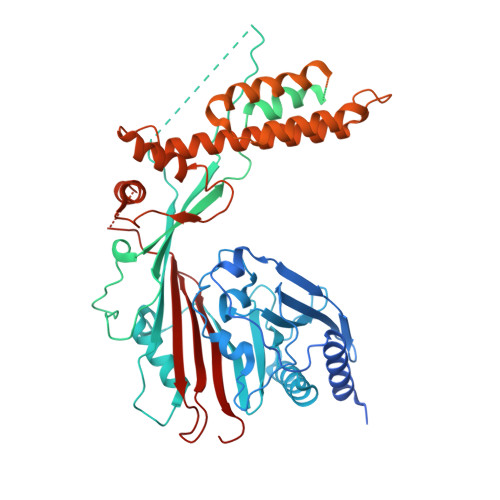
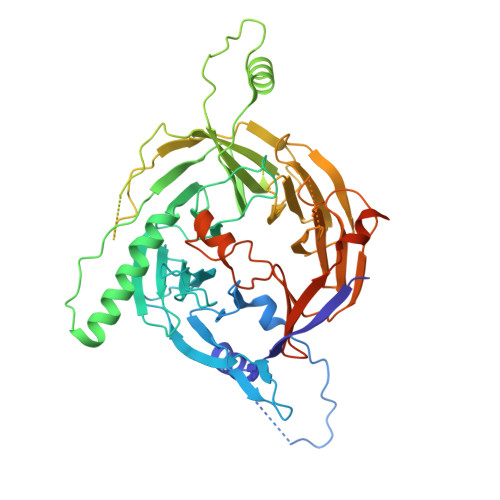
The heterodimeric Rab3GAP complex is a guanine nucleotide exchange factor (GEF) for the Rab18 GTPase that regulates lipid droplet metabolism, ER-to-Golgi trafficking, secretion, and autophagy. Why both subunits of Rab3GAP are required for Rab18 GEF activity and the molecular basis of how Rab3GAP engages and activates its cognate substrate are unknown. Here we show that human Rab3GAP is conformationally flexible and potentially autoinhibited by the C-terminal domain of its Rab3GAP2 subunit. Our high-resolution structure of the catalytic core of Rab3GAP, determined by cryo-EM, shows that the Rab3GAP2 N-terminal domain binds Rab3GAP1 via an extensive interface. AlphaFold3 modelling analysis together with targeted mutagenesis and in vitro activity assay reveal that Rab3GAP likely engages its substrate Rab18 through an interface away from the switch and interswitch regions. Lastly, we find that three Warburg Micro Syndrome-associated missense mutations do not affect the overall architecture of Rab3GAP but instead likely interfere with substrate binding.
- Life Sciences Institute, Department of Biochemistry and Molecular Biology, The University of British Columbia, Vancouver, BC, V6T 1Z3, Canada.
Organizational Affiliation: