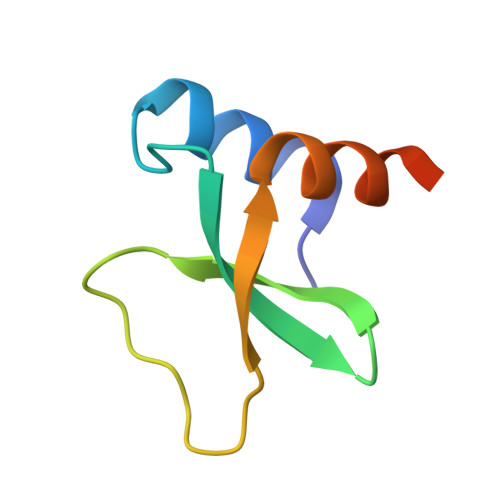
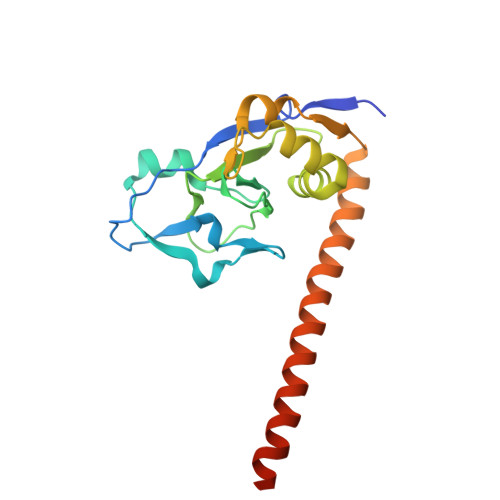
The phage protein paratox is a multifunctional metabolic regulator of Streptococcus.
Muna, T.H., Rutbeek, N.R., Horne, J., Lao, Y.W., Krokhin, O.V., Prehna, G.(2025) Nucleic Acids Res 53
- PubMed: 39673798
- DOI: https://doi.org/10.1093/nar/gkae1200
- Primary Citation of Related Structures:
8VSQ, 8VSR - PubMed Abstract:
Streptococcus pyogenes, or Group A Streptococcus (GAS), is a commensal bacteria and human pathogen. Central to GAS pathogenesis is the presence of prophage encoded virulence genes. The conserved phage gene for the protein paratox (Prx) is genetically linked to virulence genes, but the reason for this linkage is unknown. Prx inhibits GAS quorum sensing and natural competence by binding the transcription factor ComR. However, inhibiting ComR does not explain the virulence gene linkage. To address this, we took a mass spectrometry approach to search for other Prx interaction partners. The data demonstrates that Prx binds numerous DNA-binding proteins and transcriptional regulators. We show binding of Prx in vitro with the GAS protein Esub1 (SpyM3_0890) and the phage protein JM3 (SpyM3_1246). An Esub1:Prx complex X-ray crystal structure reveals that Esub1 and ComR possess a conserved Prx-binding helix. Computational modelling predicts that the Prx-binding helix is present in several, but not all, binding partners. Namely, JM3 lacks the Prx-binding helix. As Prx is conformationally dynamic, this suggests partner-dependent binding modes. Overall, Prx acts as a metabolic regulator of GAS to maintain the phage genome. As such, Prx maybe a direct contributor to the pathogenic conversion of GAS.
- Department of Microbiology, University of Manitoba, 45 Chancellors Circle, Buller Building, Winnipeg MB, R3T 2N2, Canada.
Organizational Affiliation: