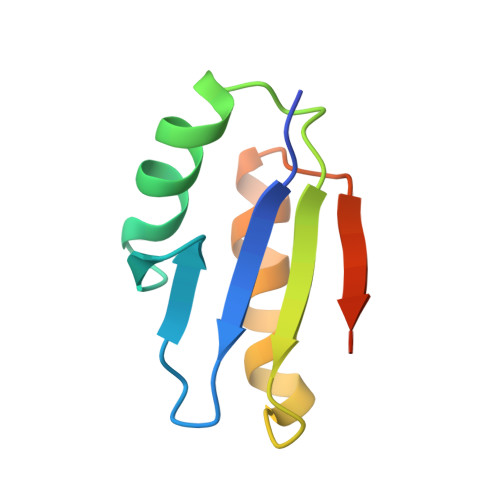
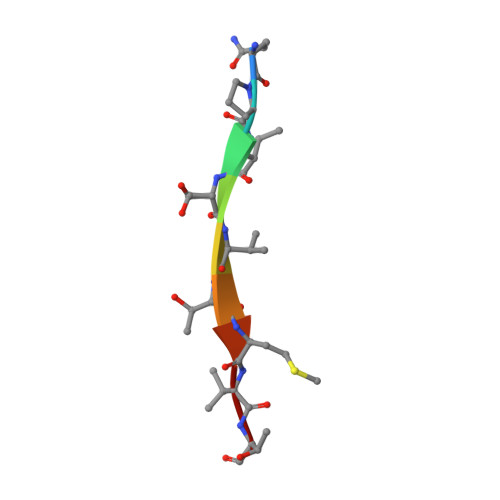
Discovery and structural characterization of the D-box, a conserved TonB motif that couples an inner-membrane motor to outer-membrane transport.
Loll, P.J., Grasty, K.C., Shultis, D.D., Guzman, N.J., Wiener, M.C.(2024) J Biological Chem 300: 105723-105723
- PubMed: 38311172
- DOI: https://doi.org/10.1016/j.jbc.2024.105723
- Primary Citation of Related Structures:
8VGC, 8VGD - PubMed Abstract:
Gram-negative bacteria use TonB-dependent transport to take up nutrients from the external environment, employing the Ton complex to import a variety of nutrients that are either scarce or too large to cross the outer membrane unaided. The Ton complex contains an inner-membrane motor (ExbBD) that generates force, as well as nutrient-specific transport proteins on the outer membrane. These two components are coupled by TonB, which transmits the force from the inner to the outer membrane. TonB contains an N-terminus anchored in the inner membrane, a C-terminal domain that binds the outer-membrane transporter, and a proline-rich linker connecting the two. While much is known about the interaction between TonB and outer-membrane transporters, the critical interface between TonB and ExbBD is less well understood. Here, we identify a conserved motif within TonB that we term the D-box, which serves as an attachment point for ExbD. We characterize the interaction between ExbD and the D-box both functionally and structurally, showing that a homodimer of ExbD captures one copy of the D-box peptide via beta-strand recruitment. We additionally show that both the D-box motif and ExbD are conserved in a range of Gram-negative bacteria, including members of the ESKAPE group of pathogens. The ExbD:D-box interaction is likely to represent an important aspect of force transduction between the inner and outer membranes. Given that TonB-dependent transport is an important contributor to virulence, this interaction is an intriguing potential target for novel antibacterial therapies.
- Department of Biochemistry and Molecular Biology, Drexel University College of Medicine, Philadelphia, Pennsylvania, USA. Electronic address: pjl28@drexel.edu.
Organizational Affiliation: