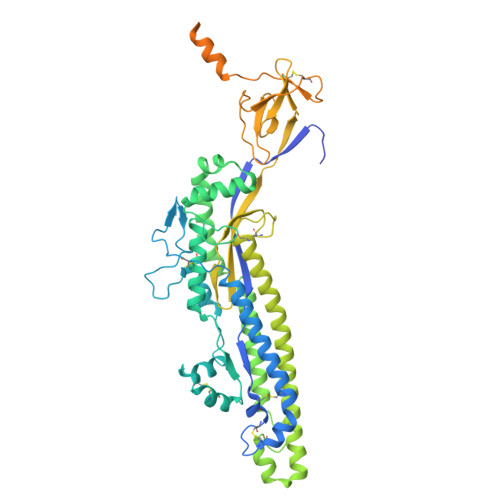
Structure of a SARS-CoV-2 spike S2 subunit in a pre-fusion, open conformation.
Olmedillas, E., Rajamanickam, R.R., Avalos, R.D., Ana-Sosa-Batiz, F., Zyla, D., Zandonatti, M.A., Harkins, S.S., Shresta, S., Hastie, K.M., Saphire, E.O.(2025) Cell Rep 44: 116052-116052
- PubMed: 40705599
- DOI: https://doi.org/10.1016/j.celrep.2025.116052
- Primary Citation of Related Structures:
8V5V - PubMed Abstract:
The continued emergence of SARS-CoV-2 variants necessitates the development of immunogens that promote broad and durable immunity. The SARS-CoV-2 S2 fusion subunit drives viral entry and has sequence conservation among coronavirus spike proteins. Therefore, S2 could represent an immunogen to boost broadly reactive antibodies. However, when expressed without the S1 domain, metastable S2 irreversibly collapses into the post-fusion six-helix bundle conformation. Beyond well-characterized RBD/NTD shifts, biophysical measurements indicate that spike exhibits reversible "breathing" motions. Using an engineered S2-only antigen that retains the pre-fusion viral surface conformation, we isolated S2-specific antibodies from convalescent and vaccinated individuals. One mAb was used to solve a high-resolution cryo-EM structure of pre-fusion S2. Our structure reveals that, relative to intact spike, engineered S2 adopts a more "open" conformation with stabilizing intermolecular interactions at the trimer base and fusion peptide repositioning. This structure could advance next-generation "booster" immunogens and illuminate potential breathing adjustments of the coronavirus spike.
- Center for Vaccine Innovation, La Jolla Institute for Immunology, La Jolla, CA 92037, USA.
Organizational Affiliation: