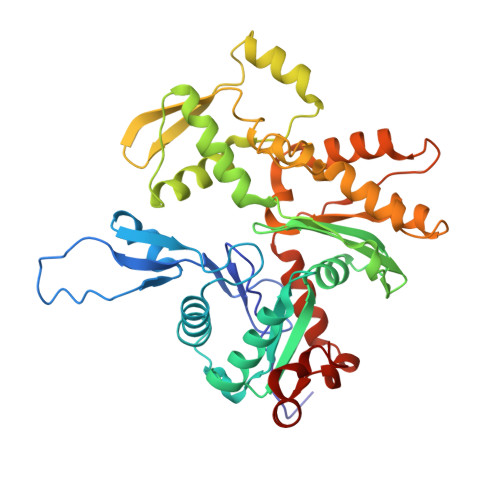
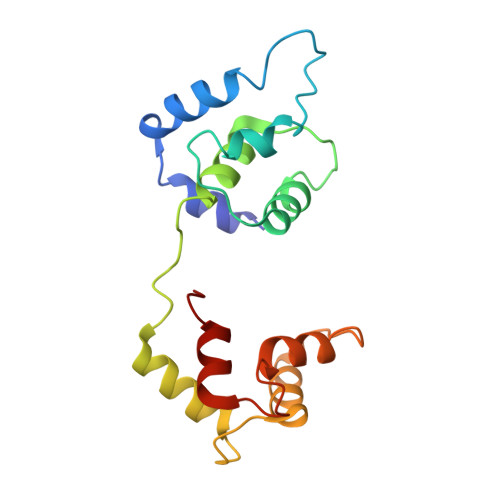
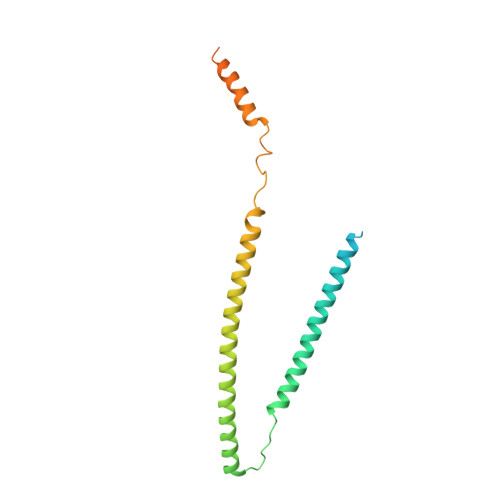
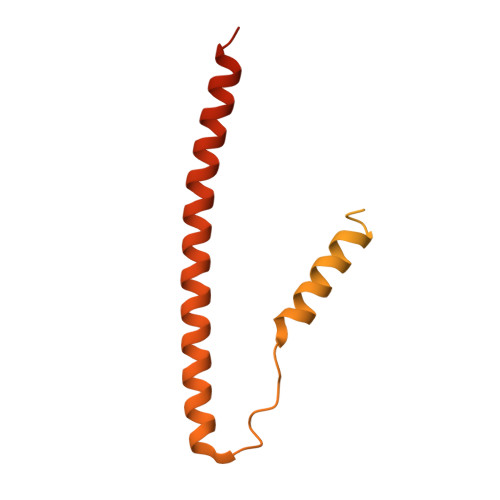
Troponin Structural Dynamics in the Native Cardiac Thin Filament Revealed by Cryo Electron Microscopy.
Risi, C.M., Belknap, B., Atherton, J., Coscarella, I.L., White, H.D., Bryant Chase, P., Pinto, J.R., Galkin, V.E.(2024) J Mol Biology 436: 168498-168498
- PubMed: 38387550
- DOI: https://doi.org/10.1016/j.jmb.2024.168498
- Primary Citation of Related Structures:
8UWW, 8UWX, 8UWY, 8UYD, 8UZ5, 8UZ6, 8UZX, 8UZY, 8V01, 8V0I, 8V0K, 8V0Y - PubMed Abstract:
Cardiac muscle contraction occurs due to repetitive interactions between myosin thick and actin thin filaments (TF) regulated by Ca 2+ levels, active cross-bridges, and cardiac myosin-binding protein C (cMyBP-C). The cardiac TF (cTF) has two nonequivalent strands, each comprised of actin, tropomyosin (Tm), and troponin (Tn). Tn shifts Tm away from myosin-binding sites on actin at elevated Ca 2+ levels to allow formation of force-producing actomyosin cross-bridges. The Tn complex is comprised of three distinct polypeptides - Ca 2+ -binding TnC, inhibitory TnI, and Tm-binding TnT. The molecular mechanism of their collective action is unresolved due to lack of comprehensive structural information on Tn region of cTF. C1 domain of cMyBP-C activates cTF in the absence of Ca 2+ to the same extent as rigor myosin. Here we used cryo-EM of native cTFs to show that cTF Tn core adopts multiple structural conformations at high and low Ca 2+ levels and that the two strands are structurally distinct. At high Ca 2+ levels, cTF is not entirely activated by Ca 2+ but exists in either partially or fully activated state. Complete dissociation of TnI C-terminus is required for full activation. In presence of cMyBP-C C1 domain, Tn core adopts a fully activated conformation, even in absence of Ca 2+ . Our data provide a structural description for the requirement of myosin to fully activate cTFs and explain increased affinity of TnC to Ca 2+ in presence of active cross-bridges. We suggest that allosteric coupling between Tn subunits and Tm is required to control actomyosin interactions.
- Department of Physiological Sciences, Eastern Virginia Medical School, Norfolk, VA 23507, USA.
Organizational Affiliation: