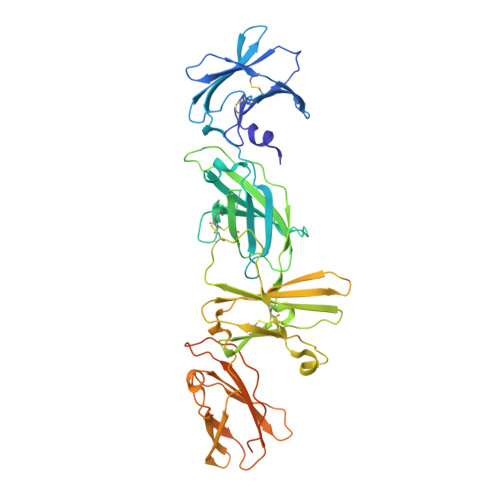
Cryo-EM structure of the extracellular domain of murine Thrombopoietin Receptor in complex with Thrombopoietin.
Sarson-Lawrence, K.T.G., Hardy, J.M., Iaria, J., Stockwell, D., Behrens, K., Saiyed, T., Tan, C., Jebeli, L., Scott, N.E., Dite, T.A., Nicola, N.A., Leis, A.P., Babon, J.J., Kershaw, N.J.(2024) Nat Commun 15: 1135-1135
- PubMed: 38326297
- DOI: https://doi.org/10.1038/s41467-024-45356-2
- Primary Citation of Related Structures:
8U18 - PubMed Abstract:
Thrombopoietin (Tpo) is the primary regulator of megakaryocyte and platelet numbers and is required for haematopoetic stem cell maintenance. Tpo functions by binding its receptor (TpoR, a homodimeric Class I cytokine receptor) and initiating cell proliferation or differentiation. Here we characterise the murine Tpo:TpoR signalling complex biochemically and structurally, using cryo-electron microscopy. Tpo uses opposing surfaces to recruit two copies of receptor, forming a 1:2 complex. Although it binds to the same, membrane-distal site on both receptor chains, it does so with significantly different affinities and its highly glycosylated C-terminal domain is not required. In one receptor chain, a large insertion, unique to TpoR, forms a partially structured loop that contacts cytokine. Tpo binding induces the juxtaposition of the two receptor chains adjacent to the cell membrane. The therapeutic agent romiplostim also targets the cytokine-binding site and the characterisation presented here supports the future development of improved TpoR agonists.
- Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, 3052, Victoria, Australia.
Organizational Affiliation: