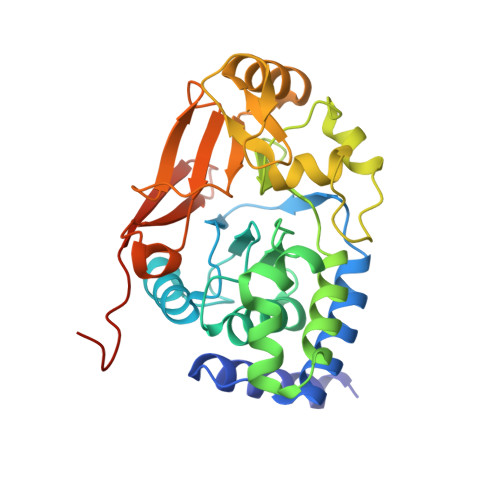
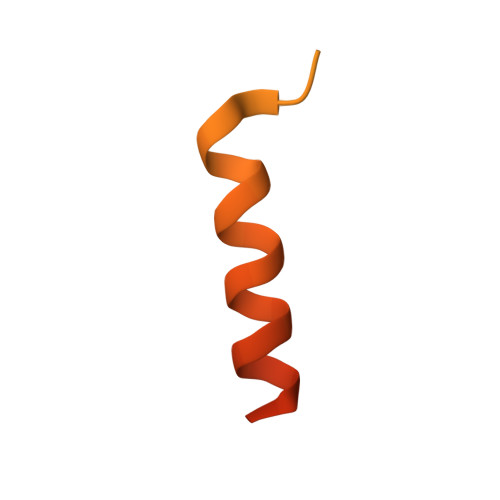
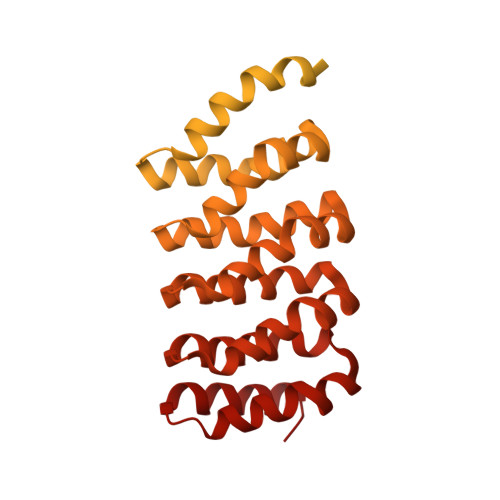Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Padi, S.K.R., Vos, M.R., Godek, R.J., Fuller, J.R., Kruse, T., Hein, J.B., Nilsson, J., Kelker, M.S., Page, R., Peti, W.(2024) Nature 625: 195-203
- PubMed: 38123684
- DOI: https://doi.org/10.1038/s41586-023-06870-3
- Primary Citation of Related Structures:
8SO0, 8TTB, 8TWE, 8TWI - PubMed Abstract:
Progression through the cell cycle is controlled by regulated and abrupt changes in phosphorylation 1 . Mitotic entry is initiated by increased phosphorylation of mitotic proteins, a process driven by kinases 2 , whereas mitotic exit is achieved by counteracting dephosphorylation, a process driven by phosphatases, especially PP2A:B55 3 . Although the role of kinases in mitotic entry is well established, recent data have shown that mitosis is only successfully initiated when the counterbalancing phosphatases are also inhibited 4 . Inhibition of PP2A:B55 is achieved by the intrinsically disordered proteins ARPP19 5,6 and FAM122A 7 . Despite their critical roles in mitosis, the mechanisms by which they achieve PP2A:B55 inhibition is unknown. Here, we report the single-particle cryo-electron microscopy structures of PP2A:B55 bound to phosphorylated ARPP19 and FAM122A. Consistent with our complementary NMR spectroscopy studies, both intrinsically disordered proteins bind PP2A:B55, but do so in highly distinct manners, leveraging multiple distinct binding sites on B55. Our extensive structural, biophysical and biochemical data explain how substrates and inhibitors are recruited to PP2A:B55 and provide a molecular roadmap for the development of therapeutic interventions for PP2A:B55-related diseases.
- Department of Molecular Biology and Biophysics, University of Connecticut Health Center, Farmington, CT, USA.
Organizational Affiliation: