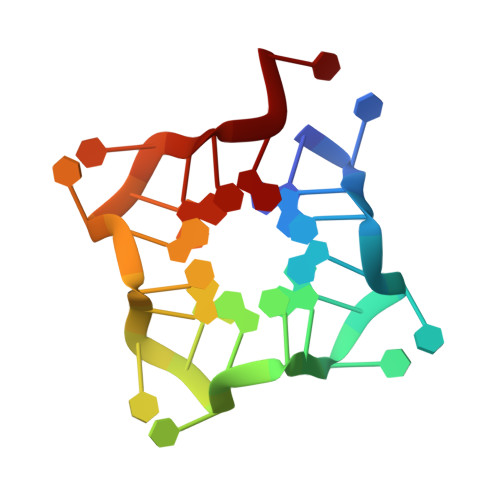
Solution Structure of Poly(UG) RNA.
Escobar, C.A., Petersen, R.J., Tonelli, M., Fan, L., Henzler-Wildman, K.A., Butcher, S.E.(2023) J Mol Biology 435: 168340-168340
- PubMed: 37924862
- DOI: https://doi.org/10.1016/j.jmb.2023.168340
- Primary Citation of Related Structures:
8TNS - PubMed Abstract:
Poly(UG) or "pUG" RNAs are UG or GU dinucleotide repeat sequences which are highly abundant in eukaryotes. Post-transcriptional addition of pUGs to RNA 3' ends marks mRNAs as vectors for gene silencing in C. elegans. We previously determined the crystal structure of pUG RNA bound to the ligand N-methyl mesoporphyrin IX (NMM), but the structure of free pUG RNA is unknown. Here we report the solution structure of the free pUG RNA (GU) 12 , as determined by nuclear magnetic resonance spectroscopy and small and wide-angle x-ray scattering (NMR-SAXS-WAXS). The low complexity sequence and 4-fold symmetry of the structure result in overlapped NMR signals that complicate chemical shift assignment. We therefore utilized single site-specific deoxyribose modifications which did not perturb the structure and introduced well-resolved methylene signals that are easily identified in NMR spectra. The solution structure ensemble has a root mean squared deviation (RMSD) of 0.62 Å and is a compact, left-handed quadruplex with a Z-form backbone, or "pUG fold." Overall, the structure agrees with the crystal structure of (GU) 12 bound to NMM, indicating the pUG fold is unaltered by docking of the NMM ligand. The solution structure reveals conformational details that could not be resolved by x-ray crystallography, which explain how the pUG fold can form within longer RNAs.
- Department of Biochemistry, University of Wisconsin-Madison, Madison, WI, USA.
Organizational Affiliation: