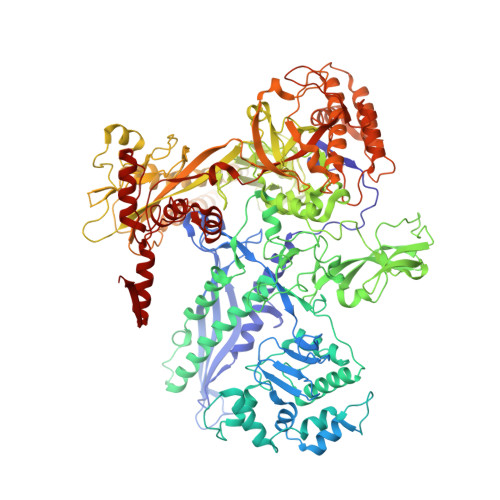
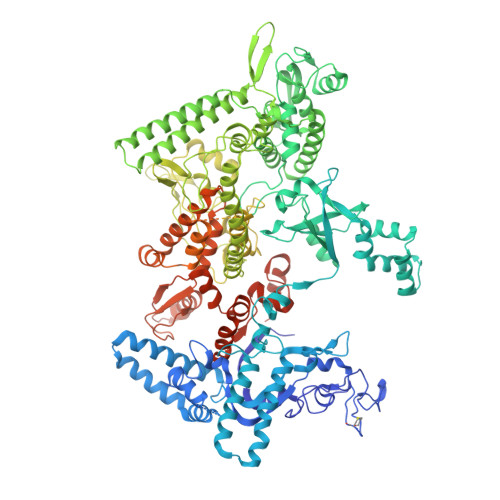
A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Oh, J., Shan, Z., Hoshika, S., Xu, J., Chong, J., Benner, S.A., Lyumkis, D., Wang, D.(2023) Nat Commun 14: 8219-8219
- PubMed: 38086811
- DOI: https://doi.org/10.1038/s41467-023-43735-9
- Primary Citation of Related Structures:
8SY5, 8SY6, 8SY7 - PubMed Abstract:
Artificially Expanded Genetic Information Systems (AEGIS) add independently replicable unnatural nucleotide pairs to the natural G:C and A:T/U pairs found in native DNA, joining the unnatural pairs through alternative modes of hydrogen bonding. Whether and how AEGIS pairs are recognized and processed by multi-subunit cellular RNA polymerases (RNAPs) remains unknown. Here, we show that E. coli RNAP selectively recognizes unnatural nucleobases in a six-letter expanded genetic system. High-resolution cryo-EM structures of three RNAP elongation complexes containing template-substrate UBPs reveal the shared principles behind the recognition of AEGIS and natural base pairs. In these structures, RNAPs are captured in an active state, poised to perform the chemistry step. At this point, the unnatural base pair adopts a Watson-Crick geometry, and the trigger loop is folded into an active conformation, indicating that the mechanistic principles underlying recognition and incorporation of natural base pairs also apply to AEGIS unnatural base pairs. These data validate the design philosophy of AEGIS unnatural basepairs. Further, we provide structural evidence supporting a long-standing hypothesis that pair mismatch during transcription occurs via tautomerization. Together, our work highlights the importance of Watson-Crick complementarity underlying the design principles of AEGIS base pair recognition.
- Division of Pharmaceutical Sciences, Skaggs School of Pharmacy & Pharmaceutical Sciences, University of California, San Diego, La Jolla, CA, 92093, USA.
Organizational Affiliation: