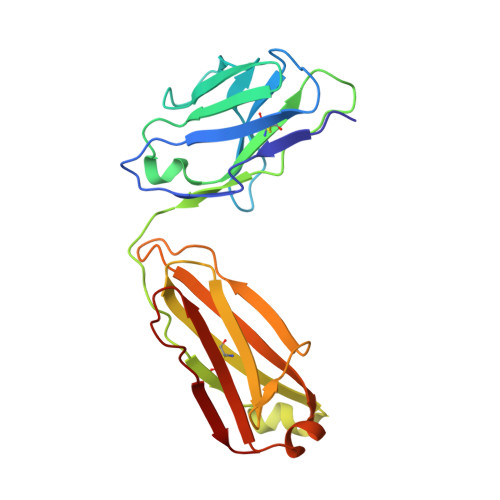
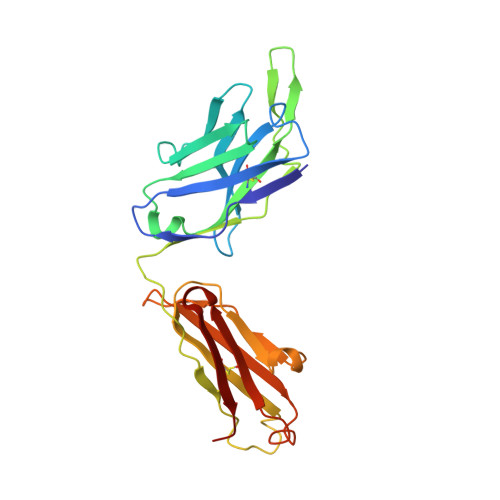
Synthetic development of a broadly neutralizing antibody against snake venom long-chain neurotoxins
Khalek, I.S., Senji Laxme, R.R., Nguyen, Y.T.K., Khochare, S., Patel, R.N., Woehl, J., Smith, J.M., Saye-Francisco, K., Kim, Y., Misson Mindrebo, L., Tran, Q., Kedzior, M., Bore, E., Limbo, O., Verma, M., Stanfield, R.L., Menzies, S.K., Ainsworth, S., Harrison, R.A., Burton, D.R., Sok, D., Wilson, I.A., Casewell, N.R., Sunagar, K., Jardine, J.G.(2024) Sci Transl Med 16: eadk1867
- PubMed: 38381847
- DOI: https://doi.org/10.1126/scitranslmed.adk1867
- Primary Citation of Related Structures:
8SXP - PubMed Abstract:
Snakebite envenoming is a major global public health concern for which improved therapies are urgently needed. The antigenic diversity present in snake venom toxins from various species presents a considerable challenge to the development of a universal antivenom. Here, we used a synthetic human antibody library to find and develop an antibody that neutralizes long-chain three-finger α-neurotoxins produced by numerous medically relevant snakes. Our antibody bound diverse toxin variants with high affinity, blocked toxin binding to the nicotinic acetylcholine receptor in vitro, and protected mice from lethal venom challenge. Structural analysis of the antibody-toxin complex revealed a binding mode that mimics the receptor-toxin interaction. The overall workflow presented is generalizable for the development of antibodies that target conserved epitopes among antigenically diverse targets, and it offers a promising framework for the creation of a monoclonal antibody-based universal antivenom to treat snakebite envenoming.
- Department of Immunology and Microbiology, Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: