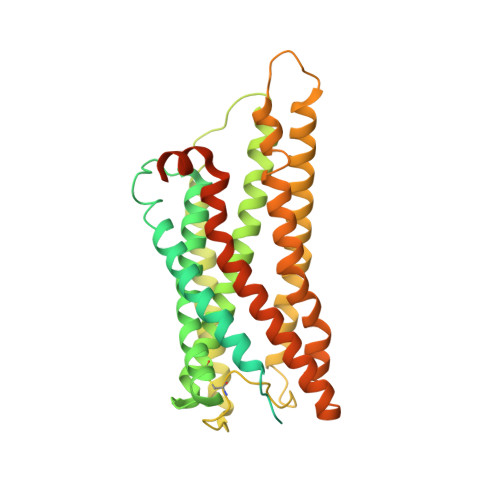
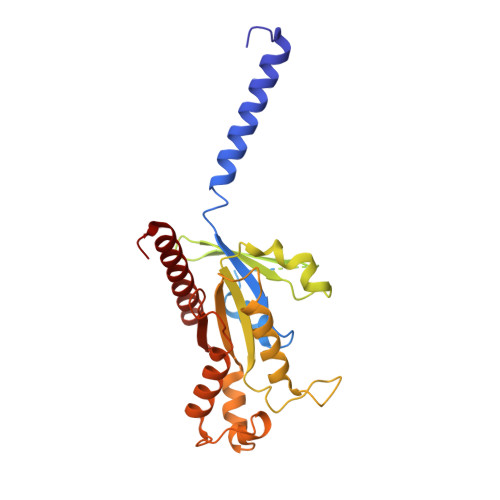
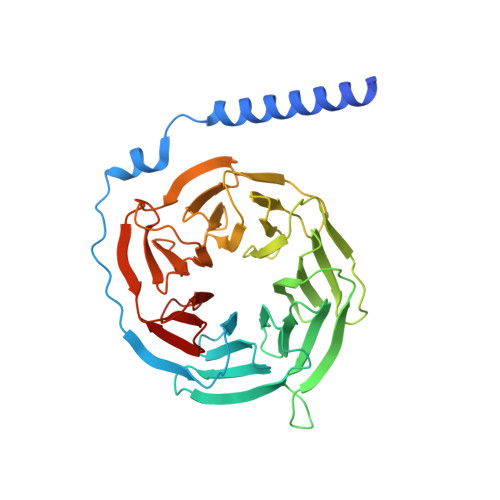
Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Xia, A., Yong, X., Zhang, C., Lin, G., Jia, G., Zhao, C., Wang, X., Hao, Y., Wang, Y., Zhou, P., Yang, X., Deng, Y., Wu, C., Chen, Y., Zhu, J., Tang, X., Liu, J., Zhang, S., Zhang, J., Xu, Z., Hu, Q., Zhao, J., Yue, Y., Yan, W., Su, Z., Wei, Y., Zhou, R., Dong, H., Shao, Z., Yang, S.(2023) Proc Natl Acad Sci U S A 120: e2308435120-e2308435120
- PubMed: 37733739
- DOI: https://doi.org/10.1073/pnas.2308435120
- Primary Citation of Related Structures:
8IYX, 8SAI - PubMed Abstract:
GPR34 is a functional G-protein-coupled receptor of Lysophosphatidylserine (LysoPS), and has pathogenic roles in numerous diseases, yet remains poorly targeted. We herein report a cryo-electron microscopy (cryo-EM) structure of GPR34 bound with LysoPS (18:1) and G i protein, revealing a unique ligand recognition mode with the negatively charged head group of LysoPS occupying a polar cavity formed by TM3, 6 and 7, and the hydrophobic tail of LysoPS residing in a lateral open hydrophobic groove formed by TM3-5. Virtual screening and subsequent structural optimization led to the identification of a highly potent and selective antagonist (YL-365). Design of fusion proteins allowed successful determination of the challenging cryo-EM structure of the inactive GPR34 complexed with YL-365, which revealed the competitive binding of YL-365 in a portion of the orthosteric binding pocket of GPR34 and the antagonist-binding-induced allostery in the receptor, implicating the inhibition mechanism of YL-365. Moreover, YL-365 displayed excellent activity in a neuropathic pain model without obvious toxicity. Collectively, this study offers mechanistic insights into the endogenous agonist recognition and antagonist inhibition of GPR34, and provides proof of concept that targeting GPR34 represents a promising strategy for disease treatment.
- Department of Biotherapy, Cancer Center and Kidney Research Institute, State Key Laboratory of Biotherapy and National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University, Chengdu, Sichuan 610041, China.
Organizational Affiliation: