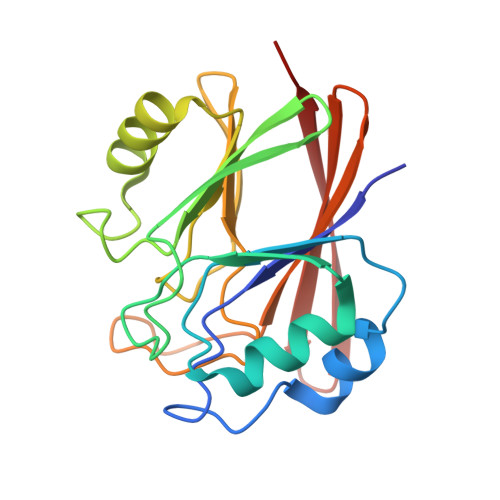
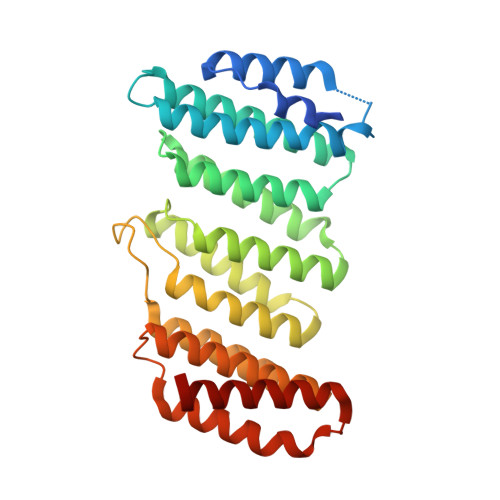
Stabilization of the retromer complex: Analysis of novel binding sites of bis-1,3-phenyl guanylhydrazone 2a to the VPS29/VPS35 interface.
Fagnani, E., Boni, F., Seneci, P., Gornati, D., Muzio, L., Mastrangelo, E., Milani, M.(2024) Comput Struct Biotechnol J 23: 1088-1093
- PubMed: 38487369
- DOI: https://doi.org/10.1016/j.csbj.2024.02.026
- Primary Citation of Related Structures:
8R02, 8R0J - PubMed Abstract:
The stabilization of the retromer protein complex can be effective in the treatment of different neurological disorders. Following the identification of bis-1,3-phenyl guanylhydrazone 2a as an effective new compound for the treatment of amyotrophic lateral sclerosis, in this work we analyze the possible binding sites of this molecule to the VPS35/VPS29 dimer of the retromer complex. Our results show that the affinity for different sites of the protein assembly depends on compound charge and therefore slight changes in the cell microenvironment could promote different binding states. Finally, we describe a novel binding site located in a deep cleft between VPS29 and VPS35 that should be further explored to select novel molecular chaperones for the stabilization of the retromer complex.
- Biophysics Institute, CNR-IBF, Via Corti 12, I-20133 Milano, Italy.
Organizational Affiliation: