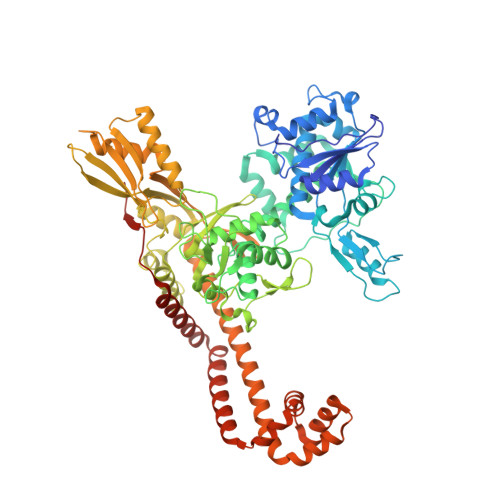
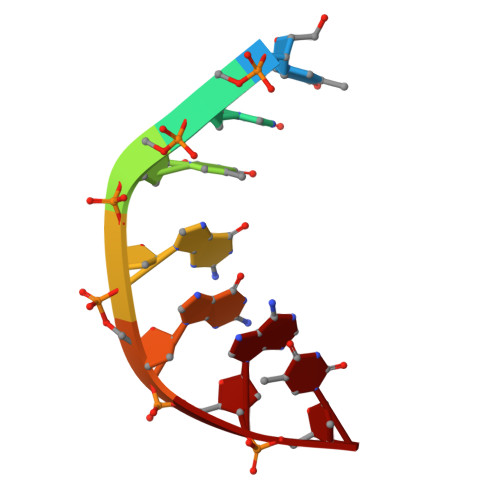
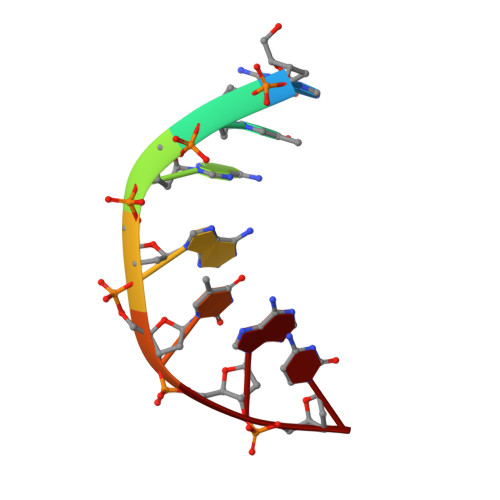
Structural basis of topoisomerase targeting by delafloxacin.
Najmudin, S., Pan, X.S., Wang, B., Govada, L., Chayen, N.E., Rubio, N., Shaffer, M.S.P., Rzepa, H.S., Fisher, L.M., Sanderson, M.R.(2025) Nat Commun 16: 5829-5829
- PubMed: 40595483
- DOI: https://doi.org/10.1038/s41467-025-60688-3
- Primary Citation of Related Structures:
8C41, 8QMB, 8QMC - PubMed Abstract:
Delafloxacin is a potent anionic fluoroquinolone approved for the treatment of respiratory infections that acts by trapping the DNA cleavage complexes of bacterial topoisomerase IV and gyrase. Its N-1-pyridinyl-, C-7-azetidinyl- and C-8-chlorine substituents confer enhanced antibiotic activity against bacteria resistant to other fluoroquinolones, but its mode of action is unclear. Here we present the X-ray crystal structures of a delafloxacin-DNA cleavage complex obtained by co-crystallization with Streptococcus pneumoniae topo IV using a graphene nucleant and solved at 2.0 and 2.4 Å resolution. The two Mg 2+ -chelated delafloxacin molecules intercalated at the DNA cleavage site are bound in an unusual conformation involving interacting out-of-plane N-1-aromatic- and C-8-chlorine- substituents. The unprecedented resolution allows comprehensive imaging of water-metal ion links integrating enzyme and DNA through drug-bound and active-site Mg 2+ ions plus the discovery of enzyme-bound K + ions. Our studies on delafloxacin action suggest that intrinsic target affinity contributes to its activity against quinolone-resistant bacteria.
- Molecular and Cellular Sciences Section, Neuroscience and Cell Biology Research Institute, City St George's, University of London, Cranmer Terrace, London, UK.
Organizational Affiliation: