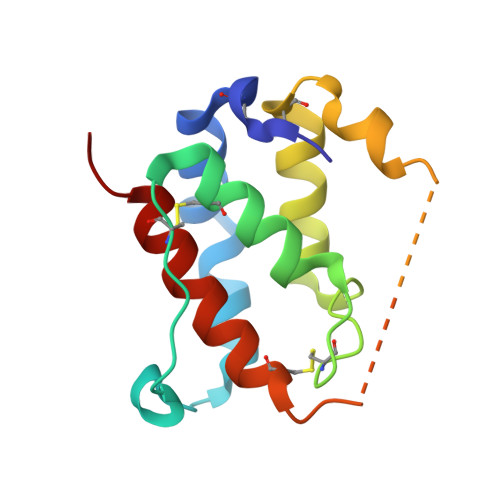
Discovery and Characterization of a Bicyclic Peptide (Bicycle) Binder to Thymic Stromal Lymphopoietin.
Narjes, F., Edfeldt, F., Petersen, J., Oster, L., Hamblet, C., Bird, J., Bold, P., Rae, R., Back, E., Stomilovic, S., Zlatoidsky, P., Svensson, T., Hidestal, L., Kunalingam, L., Shamovsky, I., De Maria, L., Gordon, E., Lewis, R.J., Watcham, S., van Rietschoten, K., Mudd, G.E., Harrison, H., Chen, L., Skynner, M.J.(2024) J Med Chem 67: 2220-2235
- PubMed: 38284169
- DOI: https://doi.org/10.1021/acs.jmedchem.3c02163
- Primary Citation of Related Structures:
8QFZ - PubMed Abstract:
Thymic stromal lymphopoietin (TSLP) is an epithelial-derived pro-inflammatory cytokine involved in the development of asthma and other atopic diseases. We used Bicycle Therapeutics' proprietary phage display platform to identify bicyclic peptides (Bicycles) with high affinity for TSLP, a target that is difficult to drug with conventional small molecules due to the extended protein-protein interactions it forms with both receptors. The hit series was shown to bind to TSLP in a hotspot, that is also used by IL-7Rα. Guided by the first X-ray crystal structure of a small peptide binding to TSLP and the identification of key metabolites, we were able to improve the proteolytic stability of this series in lung S9 fractions without sacrificing binding affinity. This resulted in the potent Bicycle 46 with nanomolar affinity to TSLP ( K D = 13 nM), low plasma clearance of 6.4 mL/min/kg, and an effective half-life of 46 min after intravenous dosing to rats.
- BicycleTx Limited, Portway Building, Granta Park, Cambridge CB21 6GS, U.K.
Organizational Affiliation: