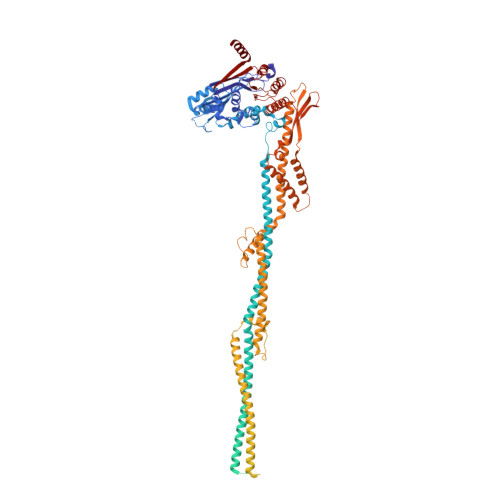
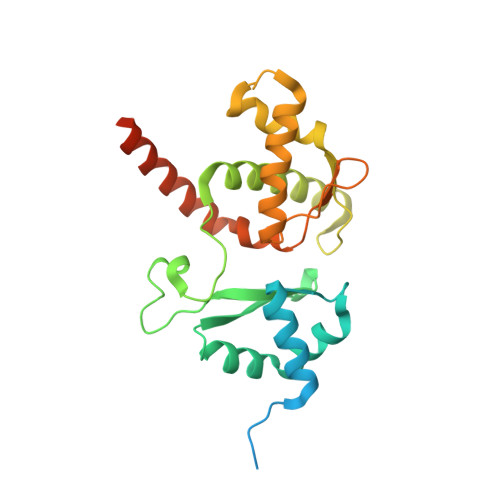
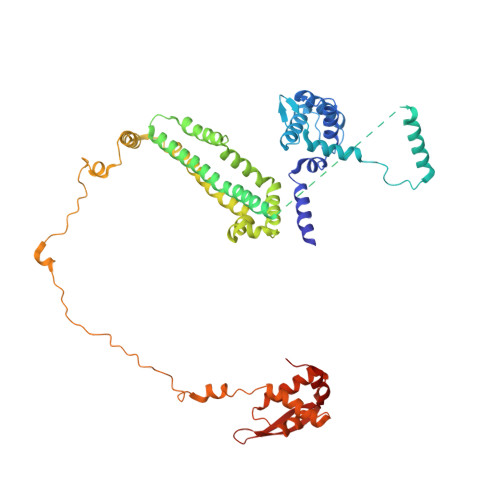
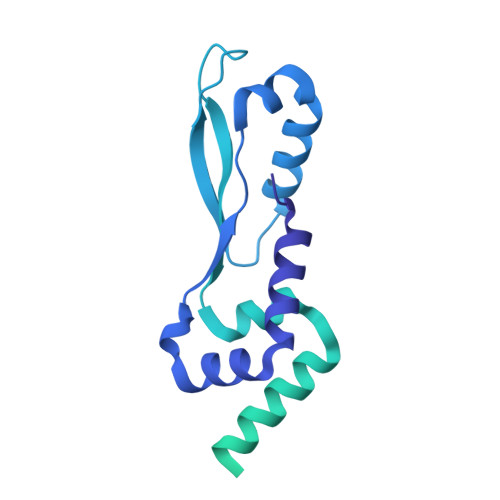
Structural basis for plasmid restriction by SMC JET nuclease.
Roisne-Hamelin, F., Liu, H.W., Taschner, M., Li, Y., Gruber, S.(2024) Mol Cell 84: 883
- PubMed: 38309275
- DOI: https://doi.org/10.1016/j.molcel.2024.01.009
- Primary Citation of Related Structures:
8Q72 - PubMed Abstract:
DNA loop-extruding SMC complexes play crucial roles in chromosome folding and DNA immunity. Prokaryotic SMC Wadjet (JET) complexes limit the spread of plasmids through DNA cleavage, yet the mechanisms for plasmid recognition are unresolved. We show that artificial DNA circularization renders linear DNA susceptible to JET nuclease cleavage. Unlike free DNA, JET cleaves immobilized plasmid DNA at a specific site, the plasmid-anchoring point, showing that the anchor hinders DNA extrusion but not DNA cleavage. Structures of plasmid-bound JetABC reveal two presumably stalled SMC motor units that are drastically rearranged from the resting state, together entrapping a U-shaped DNA segment, which is further converted to kinked V-shaped cleavage substrate by JetD nuclease binding. Our findings uncover mechanical bending of residual unextruded DNA as molecular signature for plasmid recognition and non-self DNA elimination. We moreover elucidate key elements of SMC loop extrusion, including the motor direction and the structure of a DNA-holding state.
- Department of Fundamental Microbiology (DMF), Faculty of Biology and Medicine (FBM), University of Lausanne (UNIL), 1015 Lausanne, Switzerland.
Organizational Affiliation: