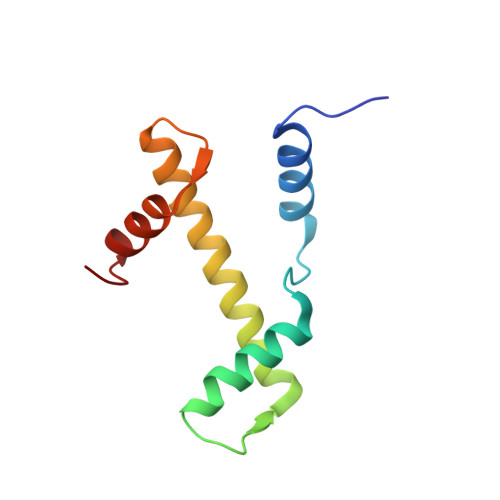
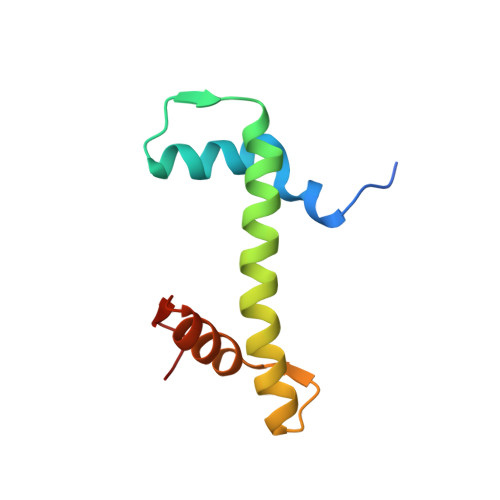
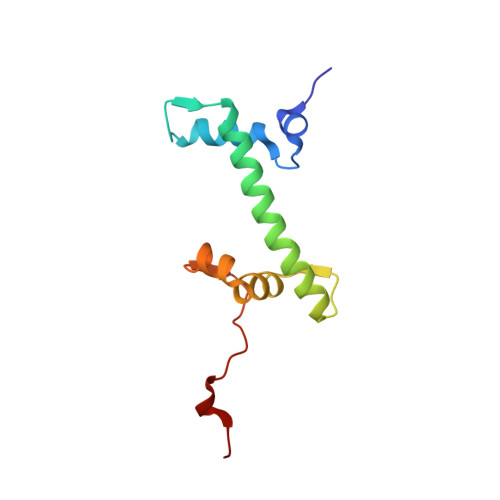
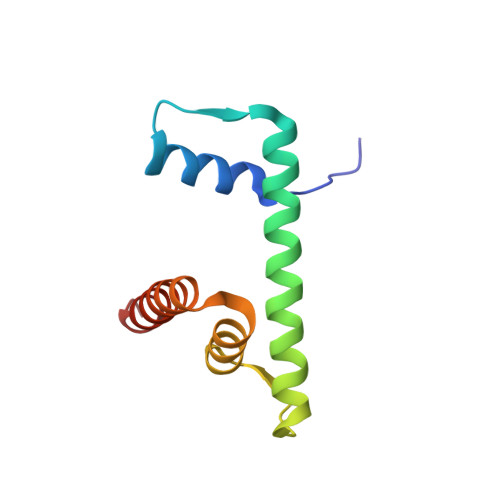
Viral peptide conjugates for metal-warhead delivery to chromatin.
Batchelor, L.K., De Falco, L., Dyson, P.J., Davey, C.A.(2024) RSC Adv 14: 8718-8725
- PubMed: 38495982
- DOI: https://doi.org/10.1039/d4ra01617c
- Primary Citation of Related Structures:
8Q36, 8Q3E, 8Q3M, 8Q3X - PubMed Abstract:
The presence of heavy metal groups can endow compounds with unique structural and chemical attributes beneficial for developing highly potent therapeutic agents and effective molecular labels. However, metallocompound binding site specificity is a major challenge that dictates the level of off-site targeting, which is a limiting factor in finding safer and more effective metal-based drugs. Here we designed and tested a family of metallopeptide conjugates based on two different chromatin-tethering viral proteins and a drug being repurposed for cancer, the Au(i) anti-arthritic auranofin. The viral peptides associate with the acidic patch of the nucleosome while the gold moiety can bind allosterically to the H3 H113 imidazole. To achieve synthesis of the conjugates, we also engineered a sulfur-free, nucleosome-binding Kaposi's sarcoma herpesvirus LANA peptide with a methionine-to-ornithine substitution and coupled the peptide to the metal group in a final step using click chemistry. The four conjugates tested are all selectively cytotoxic towards tumor cell lines, but the choice of viral peptide and mode of linkage to the Au(i) group influences metal binding site preference. Our findings suggest that viral peptide-metalloconjugates have potential for use in chromatin delivery of therapeutic warheads and as nucleosome-specific tags.
- Institut des Sciences et Ingénierie Chimiques, École Polytechnique Fédérale de Lausanne (EPFL) 1015 Lausanne Switzerland paul.dyson@epfl.ch.
Organizational Affiliation: