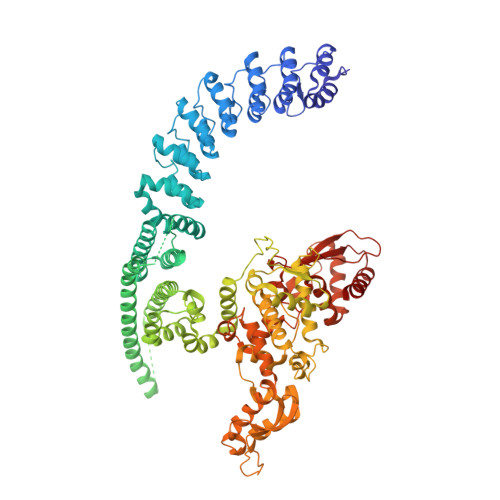
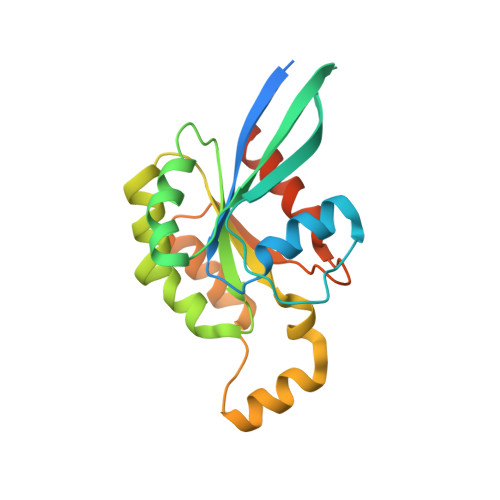
Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1.
During, J., Wolter, M., Toplak, J.J., Torres, C., Dybkov, O., Fokkens, T.J., Bohnsack, K.E., Urlaub, H., Steinchen, W., Dienemann, C., Lorenz, S.(2024) Nat Struct Mol Biol 31: 364-377
- PubMed: 38332367
- DOI: https://doi.org/10.1038/s41594-023-01203-4
- Primary Citation of Related Structures:
8PWL, 8Q0N - PubMed Abstract:
Ubiquitin ligases (E3s) are pivotal specificity determinants in the ubiquitin system by selecting substrates and decorating them with distinct ubiquitin signals. However, structure determination of the underlying, specific E3-substrate complexes has proven challenging owing to their transient nature. In particular, it is incompletely understood how members of the catalytic cysteine-driven class of HECT-type ligases (HECTs) position substrate proteins for modification. Here, we report a cryogenic electron microscopy (cryo-EM) structure of the full-length human HECT HACE1, along with solution-based conformational analyses by small-angle X-ray scattering and hydrogen-deuterium exchange mass spectrometry. Structure-based functional analyses in vitro and in cells reveal that the activity of HACE1 is stringently regulated by dimerization-induced autoinhibition. The inhibition occurs at the first step of the catalytic cycle and is thus substrate-independent. We use mechanism-based chemical crosslinking to reconstitute a complex of activated, monomeric HACE1 with its major substrate, RAC1, determine its structure by cryo-EM and validate the binding mode by solution-based analyses. Our findings explain how HACE1 achieves selectivity in ubiquitinating the active, GTP-loaded state of RAC1 and establish a framework for interpreting mutational alterations of the HACE1-RAC1 interplay in disease. More broadly, this work illuminates central unexplored aspects in the architecture, conformational dynamics, regulation and specificity of full-length HECTs.
- Research Group 'Ubiquitin Signaling Specificity', Max Planck Institute for Multidisciplinary Sciences, Göttingen, Germany.
Organizational Affiliation: