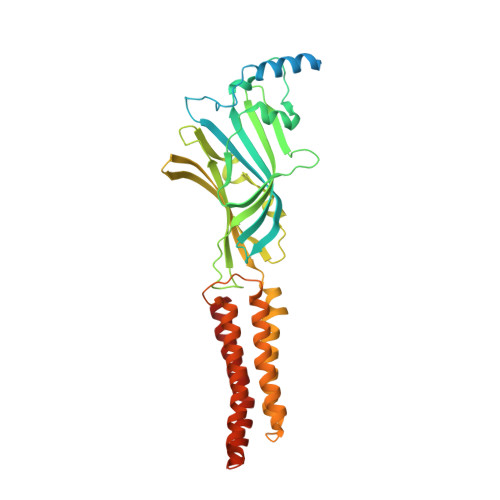
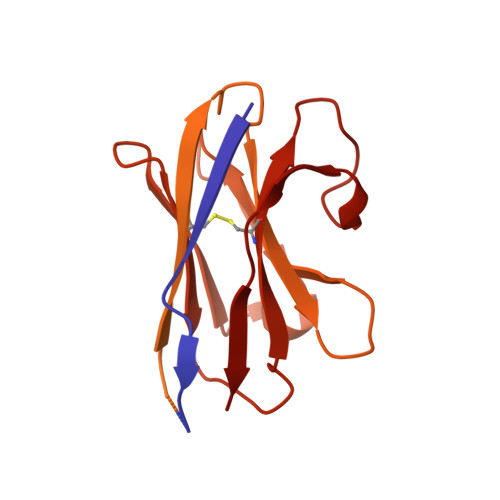
Structure determination by cryoEM at 100 keV.
McMullan, G., Naydenova, K., Mihaylov, D., Yamashita, K., Peet, M.J., Wilson, H., Dickerson, J.L., Chen, S., Cannone, G., Lee, Y., Hutchings, K.A., Gittins, O., Sobhy, M.A., Wells, T., El-Gomati, M.M., Dalby, J., Meffert, M., Schulze-Briese, C., Henderson, R., Russo, C.J.(2023) Proc Natl Acad Sci U S A 120: e2312905120-e2312905120
- PubMed: 38011573
- DOI: https://doi.org/10.1073/pnas.2312905120
- Primary Citation of Related Structures:
8PV9, 8PVA, 8PVB, 8PVC, 8PVD, 8PVE, 8PVF, 8PVG, 8PVH, 8PVI, 8PVJ - PubMed Abstract:
Electron cryomicroscopy can, in principle, determine the structures of most biological molecules but is currently limited by access, specimen preparation difficulties, and cost. We describe a purpose-built instrument operating at 100 keV-including advances in electron optics, detection, and processing-that makes structure determination fast and simple at a fraction of current costs. The instrument attains its theoretical performance limits, allowing atomic resolution imaging of gold test specimens and biological molecular structure determination in hours. We demonstrate its capabilities by determining the structures of eleven different specimens, ranging in size from 140 kDa to 2 MDa, using a fraction of the data normally required. CryoEM with a microscope designed specifically for high-efficiency, on-the-spot imaging of biological molecules will expand structural biology to a wide range of previously intractable problems.
- Medical Research Council (MRC) Laboratory of Molecular Biology, Cambridge CB2 0QH, United Kingdom.
Organizational Affiliation: