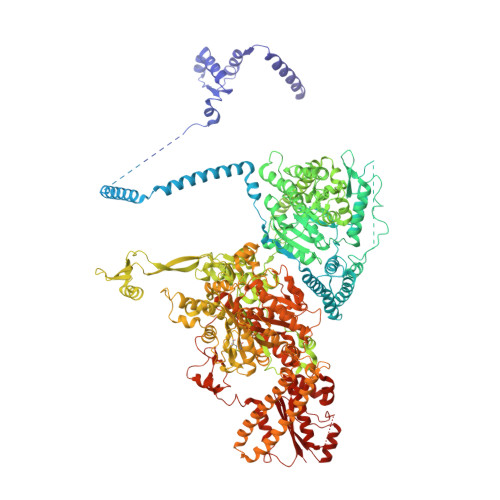
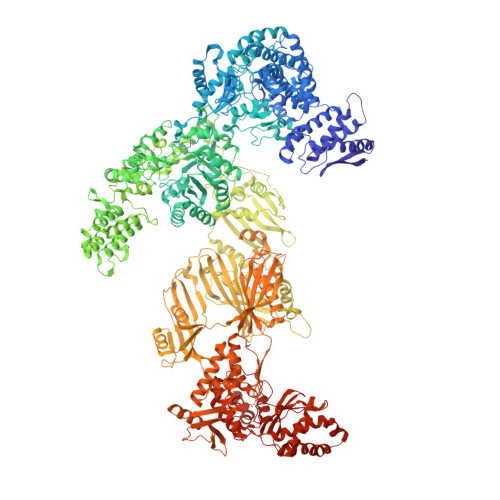
Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Singh, K., Bunzel, G., Graf, B., Yip, K.M., Neumann-Schaal, M., Stark, H., Chari, A.(2023) Cell 186: 5054-5067.e16
- PubMed: 37949058
- DOI: https://doi.org/10.1016/j.cell.2023.10.009
- Primary Citation of Related Structures:
8PRV, 8PRW, 8PS1, 8PS2, 8PS8, 8PS9, 8PSA, 8PSF, 8PSG, 8PSJ, 8PSK, 8PSL, 8PSM, 8PSP - PubMed Abstract:
Fatty acids (FAs) play a central metabolic role in living cells as constituents of membranes, cellular energy reserves, and second messenger precursors. A 2.6 MDa FA synthase (FAS), where the enzymatic reactions and structures are known, is responsible for FA biosynthesis in yeast. Essential in the yeast FAS catalytic cycle is the acyl carrier protein (ACP) that actively shuttles substrates, biosynthetic intermediates, and products from one active site to another. We resolve the S. cerevisiae FAS structure at 1.9 Å, elucidating cofactors and water networks involved in their recognition. Structural snapshots of ACP domains bound to various enzymatic domains allow the reconstruction of a full yeast FA biosynthesis cycle. The structural information suggests that each FAS functional unit could accommodate exogenous proteins to incorporate various enzymatic activities, and we show proof-of-concept experiments where ectopic proteins are used to modulate FAS product profiles.
- Department of Structural Dynamics, Max Planck Institute for Multidisciplinary Sciences, Am Fassberg 11, 37077 Göttingen, Germany.
Organizational Affiliation: