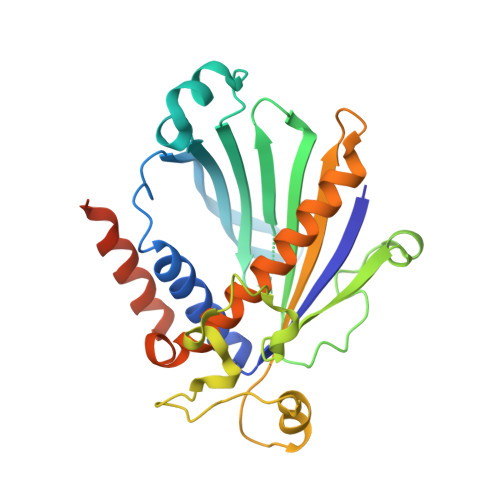
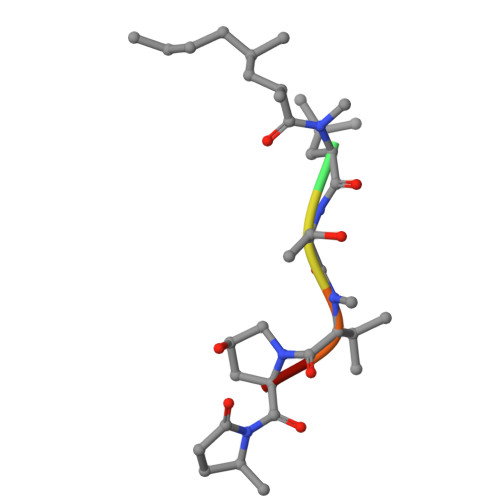
Non-vesicular phosphatidylinositol transfer plays critical roles in defining organelle lipid composition.
Kim, Y.J., Pemberton, J.G., Eisenreichova, A., Mandal, A., Koukalova, A., Rohilla, P., Sohn, M., Konradi, A.W., Tang, T.T., Boura, E., Balla, T.(2024) EMBO J 43: 2035-2061
- PubMed: 38627600
- DOI: https://doi.org/10.1038/s44318-024-00096-3
- Primary Citation of Related Structures:
8PQO - PubMed Abstract:
Phosphatidylinositol (PI) is the precursor lipid for the minor phosphoinositides (PPIns), which are critical for multiple functions in all eukaryotic cells. It is poorly understood how phosphatidylinositol, which is synthesized in the ER, reaches those membranes where PPIns are formed. Here, we used VT01454, a recently identified inhibitor of class I PI transfer proteins (PITPs), to unravel their roles in lipid metabolism, and solved the structure of inhibitor-bound PITPNA to gain insight into the mode of inhibition. We found that class I PITPs not only distribute PI for PPIns production in various organelles such as the plasma membrane (PM) and late endosomes/lysosomes, but that their inhibition also significantly reduced the levels of phosphatidylserine, di- and triacylglycerols, and other lipids, and caused prominent increases in phosphatidic acid. While VT01454 did not inhibit Golgi PI4P formation nor reduce resting PM PI(4,5)P 2 levels, the recovery of the PM pool of PI(4,5)P 2 after receptor-mediated hydrolysis required both class I and class II PITPs. Overall, these studies show that class I PITPs differentially regulate phosphoinositide pools and affect the overall cellular lipid landscape.
- Section on Molecular Signal Transduction, Eunice Kennedy Shriver, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, 20892, USA.
Organizational Affiliation: