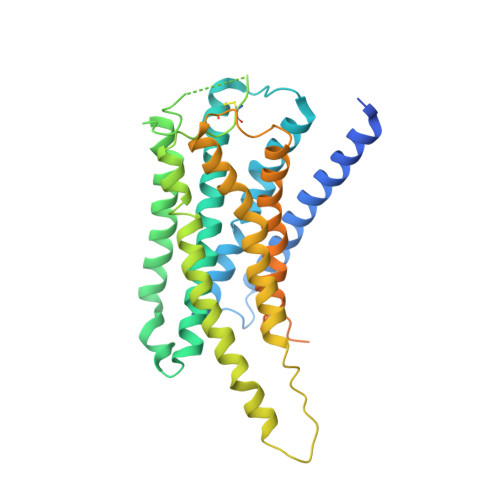
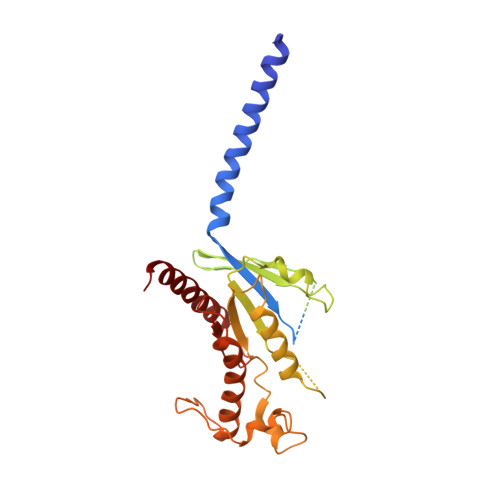
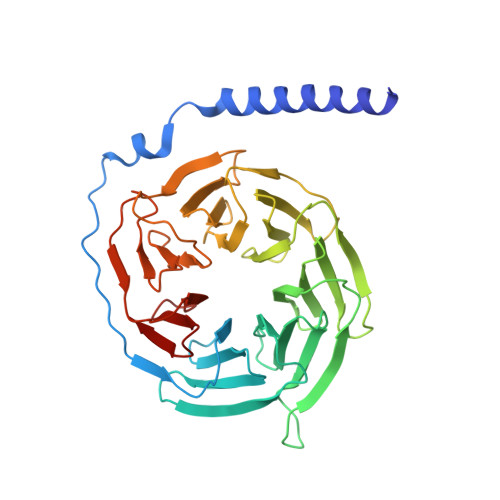
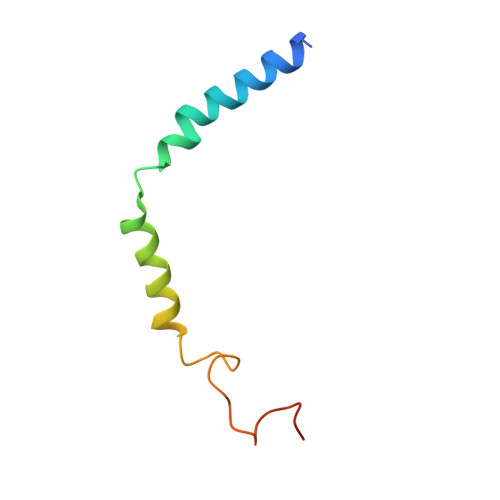
Cryo-EM structure of cell-free synthesized human histamine 2 receptor/G s complex in nanodisc environment.
Kock, Z., Schnelle, K., Persechino, M., Umbach, S., Schihada, H., Januliene, D., Parey, K., Pockes, S., Kolb, P., Dotsch, V., Moller, A., Hilger, D., Bernhard, F.(2024) Nat Commun 15: 1831-1831
- PubMed: 38418462
- DOI: https://doi.org/10.1038/s41467-024-46096-z
- Primary Citation of Related Structures:
8POK - PubMed Abstract:
Here we describe the cryo-electron microscopy structure of the human histamine 2 receptor (H 2 R) in an active conformation with bound histamine and in complex with G s heterotrimeric protein at an overall resolution of 3.4 Å. The complex was generated by cotranslational insertion of the receptor into preformed nanodisc membranes using cell-free synthesis in E. coli lysates. Structural comparison with the inactive conformation of H 2 R and the inactive and G q -coupled active state of H 1 R together with structure-guided functional experiments reveal molecular insights into the specificity of ligand binding and G protein coupling for this receptor family. We demonstrate lipid-modulated folding of cell-free synthesized H 2 R, its agonist-dependent internalization and its interaction with endogenously synthesized H 1 R and H 2 R in HEK293 cells by applying a recently developed nanotransfer technique.
- Centre for Biomolecular Magnetic Resonance, Institute for Biophysical Chemistry, Goethe-University of Frankfurt/Main, Frankfurt, Germany.
Organizational Affiliation: