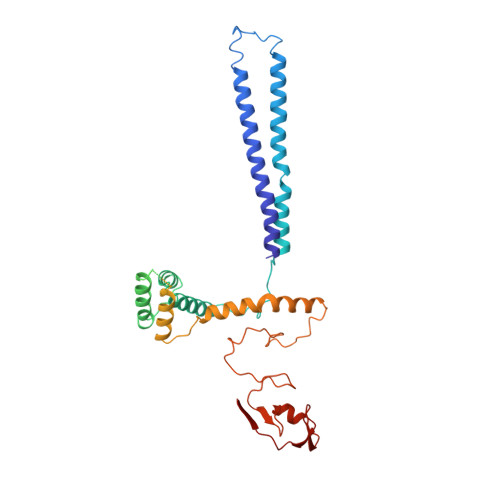
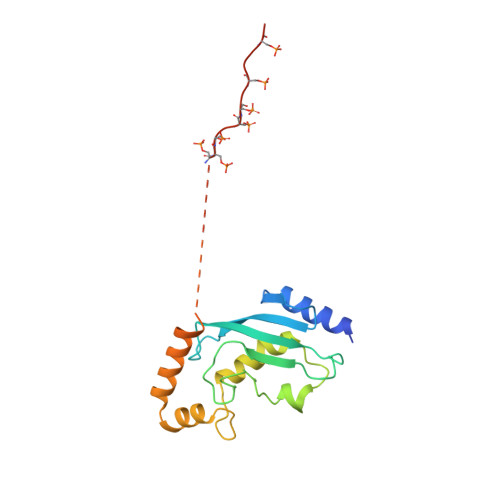
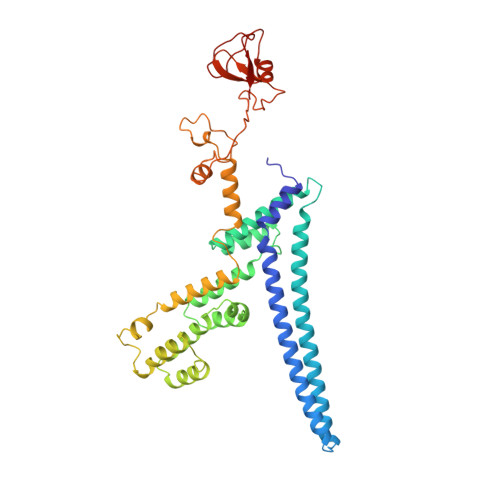
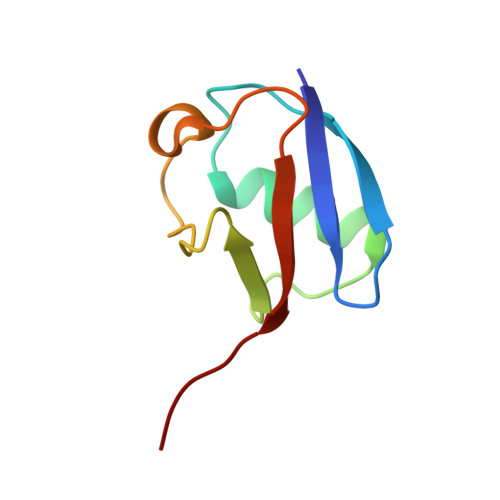
Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Chrustowicz, J., Sherpa, D., Li, J., Langlois, C.R., Papadopoulou, E.C., Vu, D.T., Hehl, L.A., Karayel, O., Beier, V., von Gronau, S., Muller, J., Prabu, J.R., Mann, M., Kleiger, G., Alpi, A.F., Schulman, B.A.(2024) Mol Cell 84: 293
- PubMed: 38113892
- DOI: https://doi.org/10.1016/j.molcel.2023.11.027
- Primary Citation of Related Structures:
8PJN, 8PMQ - PubMed Abstract:
Ubiquitylation is catalyzed by coordinated actions of E3 and E2 enzymes. Molecular principles governing many important E3-E2 partnerships remain unknown, including those for RING-family GID/CTLH E3 ubiquitin ligases and their dedicated E2, Ubc8/UBE2H (yeast/human nomenclature). GID/CTLH-Ubc8/UBE2H-mediated ubiquitylation regulates biological processes ranging from yeast metabolic signaling to human development. Here, cryoelectron microscopy (cryo-EM), biochemistry, and cell biology reveal this exquisitely specific E3-E2 pairing through an unconventional catalytic assembly and auxiliary interactions 70-100 Å away, mediated by E2 multisite phosphorylation. Rather than dynamic polyelectrostatic interactions reported for other ubiquitylation complexes, multiple Ubc8/UBE2H phosphorylation sites within acidic CK2-targeted sequences specifically anchor the E2 C termini to E3 basic patches. Positions of phospho-dependent interactions relative to the catalytic domains correlate across evolution. Overall, our data show that phosphorylation-dependent multivalency establishes a specific E3-E2 partnership, is antagonistic with dephosphorylation, rigidifies the catalytic centers within a flexing GID E3-substrate assembly, and facilitates substrate collision with ubiquitylation active sites.
- Department of Molecular Machines and Signaling, Max Planck Institute of Biochemistry, Martinsried 82152, Germany.
Organizational Affiliation: