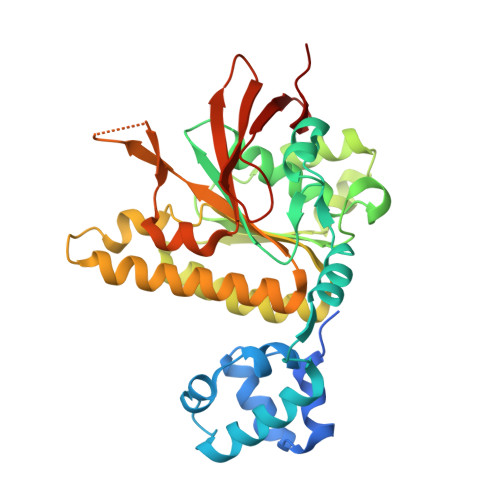
Structural basis for stabilisation of the RAD51 nucleoprotein filament by BRCA2.
Appleby, R., Joudeh, L., Cobbett, K., Pellegrini, L.(2023) Nat Commun 14: 7003-7003
- PubMed: 37919288
- DOI: https://doi.org/10.1038/s41467-023-42830-1
- Primary Citation of Related Structures:
8PBC, 8PBD - PubMed Abstract:
The BRCA2 tumour suppressor protein preserves genomic integrity via interactions with the DNA-strand exchange RAD51 protein in homology-directed repair. The RAD51-binding TR2 motif at the BRCA2 C-terminus is essential for protection and restart of stalled replication forks. Biochemical evidence shows that TR2 recognises filamentous RAD51, but existing models of TR2 binding to RAD51 lack a structural basis. Here we used cryo-electron microscopy and structure-guided mutagenesis to elucidate the mechanism of TR2 binding to nucleoprotein filaments of human RAD51. We find that TR2 binds across the protomer interface in the filament, acting as a brace for adjacent RAD51 molecules. TR2 targets an acidic-patch motif on human RAD51 that serves as a recruitment hub in fission yeast Rad51 for recombination mediators Rad52 and Rad55-Rad57. Our findings provide a structural rationale for RAD51 filament stabilisation by BRCA2 and reveal a common recruitment mechanism of recombination mediators to the RAD51 filament.
- Department of Biochemistry, University of Cambridge, Cambridge, CB2 1GA, UK.
Organizational Affiliation: