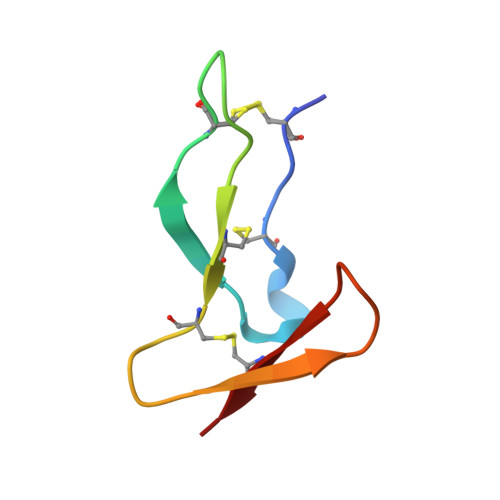
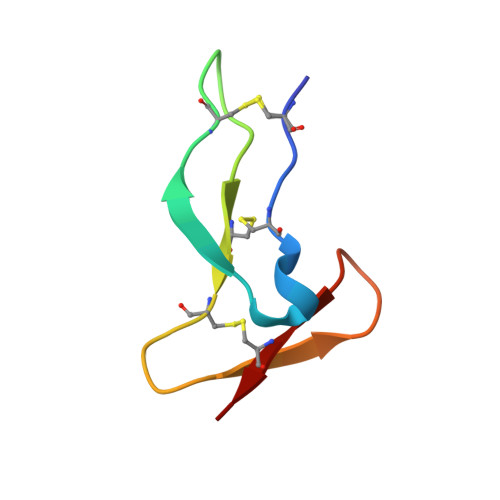
Deciphering the Synthetic and Refolding Strategy of a Cysteine-Rich Domain in the Tumor Necrosis Factor Receptor (TNF-R) for Racemic Crystallography Analysis and d-Peptide Ligand Discovery.
Lander, A.J., Kong, Y., Jin, Y., Wu, C., Luk, L.Y.P.(2024) ACS Bio Med Chem Au 4: 68-76
- PubMed: 38404743
- DOI: https://doi.org/10.1021/acsbiomedchemau.3c00060
- Primary Citation of Related Structures:
8P6Q - PubMed Abstract:
Many cell-surface receptors are promising targets for chemical synthesis because of their critical roles in disease development. This synthetic approach enables investigations by racemic protein crystallography and ligand discovery by mirror-image methodologies. However, due to their complex nature, the chemical synthesis of a receptor can be a significant challenge. Here, we describe the chemical synthesis and folding of a central, cysteine-rich domain of the cell-surface receptor tumor necrosis factor 1 which is integral to binding of the cytokine TNF-α, namely, TNFR-1 CRD2. Racemic protein crystallography at 1.4 Å confirmed that the native binding conformation was preserved, and TNFR-1 CRD2 maintained its capacity to bind to TNF-α ( K D ≈ 7 nM). Encouraged by this discovery, we carried out mirror-image phage display using the enantiomeric receptor mimic and identified a d-peptide ligand for TNFR-1 CRD2 ( K D = 1 μM). This work demonstrated that cysteine-rich domains, including the central domains, can be chemically synthesized and used as mimics for investigations.
- School of Chemistry, Cardiff University, Main Building, Park Place, Cardiff CF10 3AT, U.K.
Organizational Affiliation: