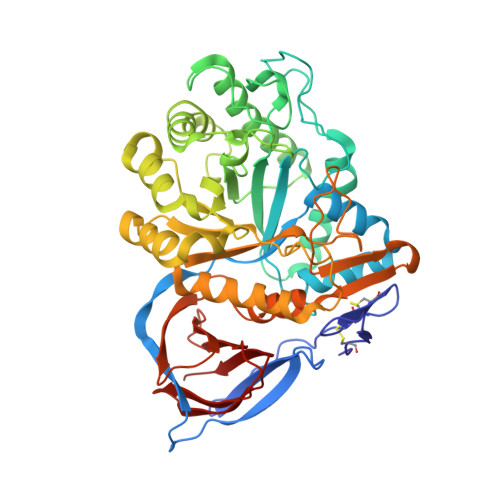
Identification of ss-Glucocerebrosidase Activators for Glucosylceramide hydrolysis.
Schulze, M.E.D., Scholz, D., Jnoff, E., Hall, A., Melin, J., Sands, Z.A., Rodriguez, E., Andre, V.M.(2024) ChemMedChem 19: e202300548-e202300548
- PubMed: 38381042
- DOI: https://doi.org/10.1002/cmdc.202300548
- Primary Citation of Related Structures:
8P3E, 8P41 - PubMed Abstract:
Several novel chemical series were identified that modulate glucocerebrosidase (GCase). Compounds from these series are active on glucosylceramide, unlike other known GCase modulators. We obtained GCase crystal structures with two compounds that have distinct chemotypes. Positive allosteric modulators bind to a site on GCase and induce conformational changes, but also induce an equilibrium state between monomer and dimer.
- UCB, 216 Bath Road, Slough, SL1 3WE, UK.
Organizational Affiliation: