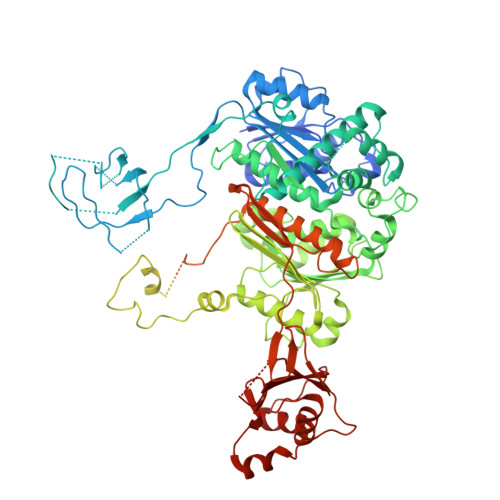
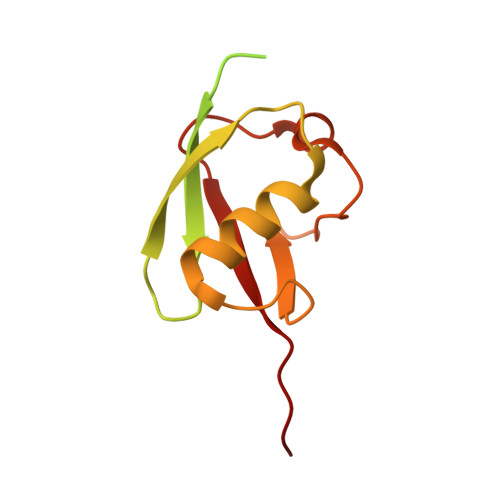
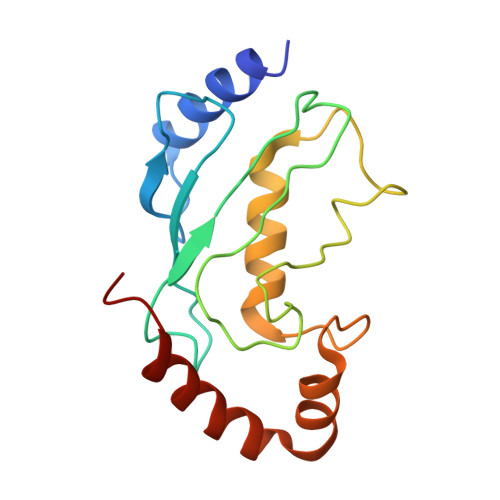
Insights into the ISG15 transfer cascade by the UBE1L activating enzyme.
Wallace, I., Baek, K., Prabu, J.R., Vollrath, R., von Gronau, S., Schulman, B.A., Swatek, K.N.(2023) Nat Commun 14: 7970-7970
- PubMed: 38042859
- DOI: https://doi.org/10.1038/s41467-023-43711-3
- Primary Citation of Related Structures:
8OIF - PubMed Abstract:
The attachment of the ubiquitin-like protein ISG15 to substrates by specific E1-E2-E3 enzymes is a well-established signalling mechanism of the innate immune response. Here, we present a 3.45 Å cryo-EM structure of a chemically trapped UBE1L-UBE2L6 complex bound to activated ISG15. This structure reveals the details of the first steps of ISG15 recognition and UBE2L6 recruitment by UBE1L (also known as UBA7). Taking advantage of viral effector proteins from severe acute respiratory coronavirus 2 (SARS-CoV-2) and influenza B virus (IBV), we validate the structure and confirm the importance of the ISG15 C-terminal ubiquitin-like domain in the adenylation reaction. Moreover, biochemical characterization of the UBE1L-ISG15 and UBE1L-UBE2L6 interactions enables the design of ISG15 and UBE2L6 mutants with altered selectively for the ISG15 and ubiquitin conjugation pathways. Together, our study helps to define the molecular basis of these interactions and the specificity determinants that ensure the fidelity of ISG15 signalling during the antiviral response.
- Medical Research Council Protein Phosphorylation and Ubiquitylation Unit, School of Life Sciences, University of Dundee, Dundee, DD1 5EH, UK.
Organizational Affiliation: