Tau-S214 Phosphorylation Inhibits Fyn Kinase Interaction and Increases the Decay Time of NMDAR-mediated Current.
Jos, S., Poulose, R., Kambaru, A., Gogoi, H., Dalavaikodihalli Nanjaiah, N., Padmanabhan, B., Mehta, B., Padavattan, S.(2024) J Mol Biology 436: 168445-168445
- PubMed: 38218365
- DOI: https://doi.org/10.1016/j.jmb.2024.168445
- Primary Citation of Related Structures:
8KDX - PubMed Abstract:
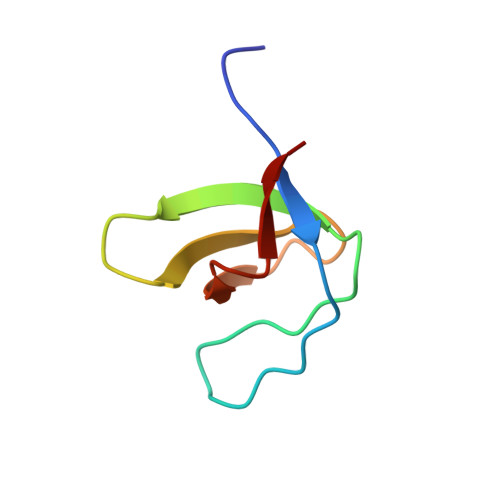
Fyn kinase SH3 domain interaction with PXXP motif in the Tau protein is implicated in AD pathology and is central to NMDAR function. Among seven PXXP motifs localized in proline-rich domain of Tau protein, tandem 5th and 6th PXXP motifs are critical to Fyn-SH3 domain interaction. Here, we report the crystal structure of Fyn-SH3 -Tau (207-221) peptide consisting of 5th and 6th PXXP motif complex to 1.01 Å resolution. Among five AD-specific phosphorylation sites encompassing the 5th and 6th PXXP motifs, only S214 residue showed interaction with SH3 domain. Biophysical studies showed that Tau (207-221) with S214-phosphorylation (pS214) inhibits its interaction with Fyn-SH3 domain. The individual administration of Tau (207-221) with/without pS214 peptides to a single neuron increased the decay time of evoked NMDA current response. Recordings of spontaneous NMDA EPSCs at +40 mV indicate an increase in frequency and amplitude of events for the Tau (207-221) peptide. Conversely, the Tau (207-221) with pS214 peptide exhibited a noteworthy amplitude increase alongside a prolonged decay time. These outcomes underscore the distinctive modalities of action associated with each peptide in the study. Overall, this study provides insights into how Tau (207-221) with/without pS214 affects the molecular framework of NMDAR signaling, indicating its involvement in Tau-related pathogenesis.
- Department of Biophysics, National Institute of Mental Health and Neurosciences, Bangalore 560029, India.
Organizational Affiliation: