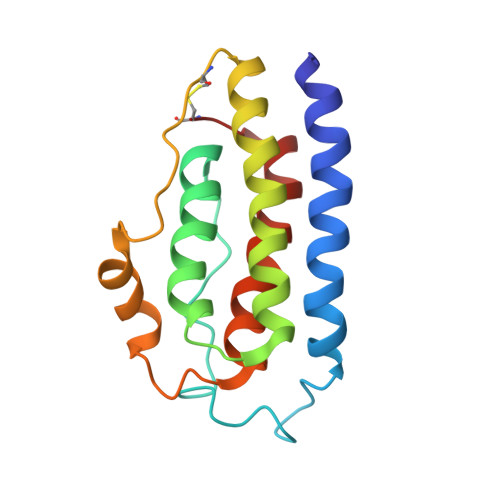
The solution structure of human leptin reveals a conformational plasticity important for receptor recognition.
Fan, X., Qin, R., Yuan, W., Fan, J.S., Huang, W., Lin, Z.(2024) Structure 32: 18-23.e2
- PubMed: 37924810
- DOI: https://doi.org/10.1016/j.str.2023.10.009
- Primary Citation of Related Structures:
8K6Z - PubMed Abstract:
Leptin is a multi-potency cytokine that regulates various physiological functions, including weight control and energy homeostasis. Signaling of leptin is also important in many aging-related diseases. Leptin is required for the noncovalent crosslinking of different extracellular domains of leptin receptors, which is critical for receptor activation and downstream signaling. Nevertheless, the structure of intact apo-form leptin and the structural transition leptin undergoes upon receptor binding are not fully understood yet. Here, we determined the monomeric structure of wild-type human leptin by solution-state nuclear magnetic resonance spectroscopy. Leptin contains an intrinsically disordered region (IDR) in the internal A-B loop and the flexible helix E in the C-D loop, both of which undergo substantial local structural changes when leptin binds to its receptor. Our findings provide further insights into the molecular mechanisms of leptin signaling.
- School of Life Sciences, Tianjin University, Tianjin 300072, China.
Organizational Affiliation: