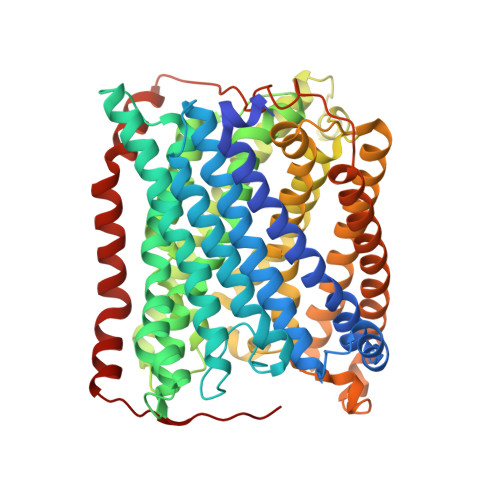
Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Safari, C., Ghosh, S., Andersson, R., Johannesson, J., Bath, P., Uwangue, O., Dahl, P., Zoric, D., Sandelin, E., Vallejos, A., Nango, E., Tanaka, R., Bosman, R., Borjesson, P., Dunevall, E., Hammarin, G., Ortolani, G., Panman, M., Tanaka, T., Yamashita, A., Arima, T., Sugahara, M., Suzuki, M., Masuda, T., Takeda, H., Yamagiwa, R., Oda, K., Fukuda, M., Tosha, T., Naitow, H., Owada, S., Tono, K., Nureki, O., Iwata, S., Neutze, R., Branden, G.(2023) Sci Adv 9: eadh4179-eadh4179
- PubMed: 38064560
- DOI: https://doi.org/10.1126/sciadv.adh4179
- Primary Citation of Related Structures:
8AJZ, 8K65, 8K6Y - PubMed Abstract:
Cytochrome c oxidase (C c O) is part of the respiratory chain and contributes to the electrochemical membrane gradient in mitochondria as well as in many bacteria, as it uses the energy released in the reduction of oxygen to pump protons across an energy-transducing biological membrane. Here, we use time-resolved serial femtosecond crystallography to study the structural response of the active site upon flash photolysis of carbon monoxide (CO) from the reduced heme a 3 of ba 3 -type C c O. In contrast with the aa 3 -type enzyme, our data show how CO is stabilized on Cu B through interactions with a transiently ordered water molecule. These results offer a structural explanation for the extended lifetime of the Cu B -CO complex in ba 3 -type C c O and, by extension, the extremely high oxygen affinity of the enzyme.
- Department of Chemistry and Molecular Biology, University of Gothenburg, Box 462, SE-40530 Gothenburg, Sweden.
Organizational Affiliation: