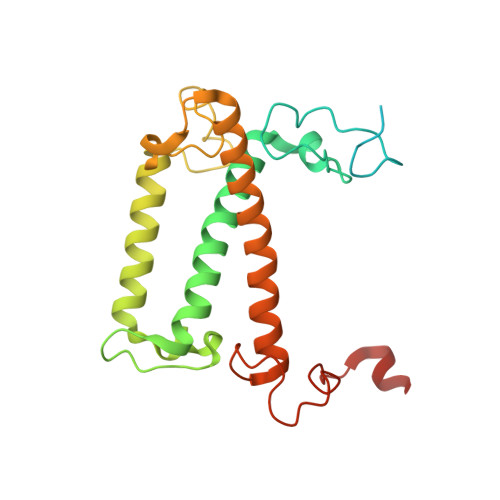
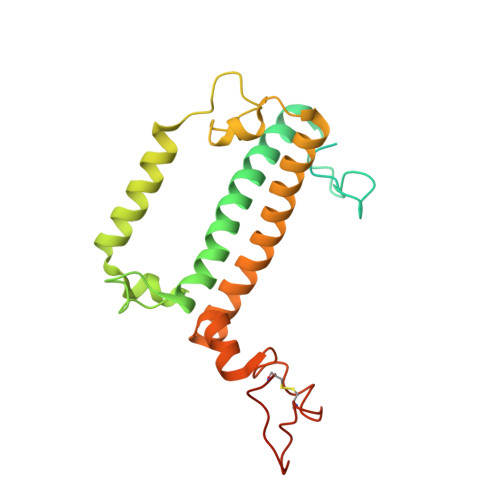
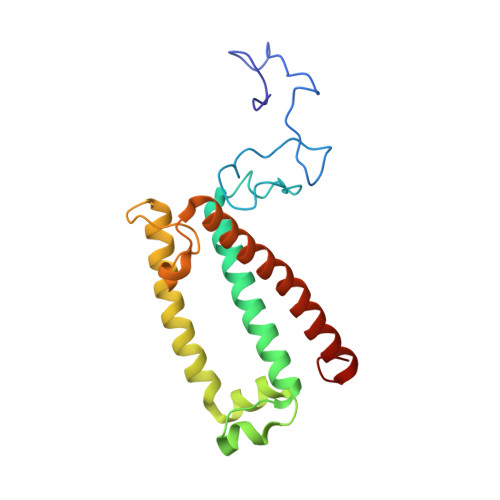
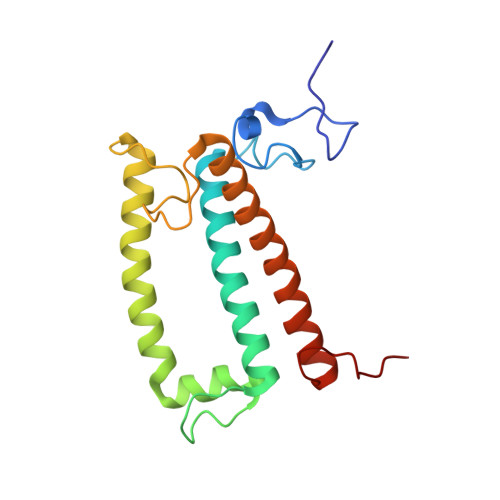
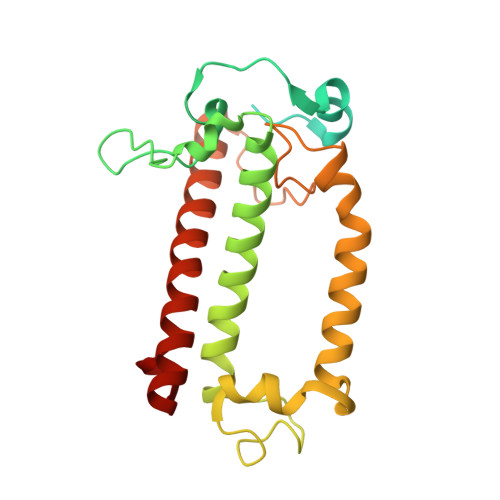
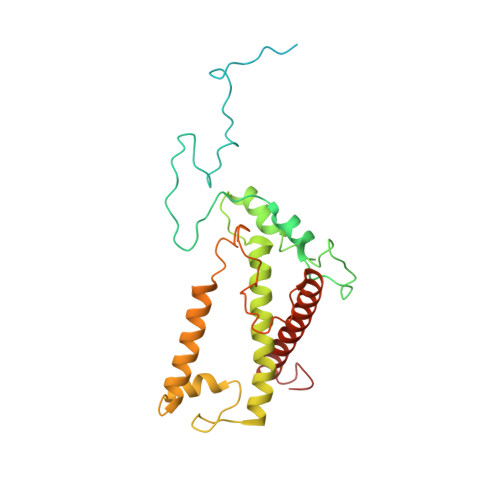
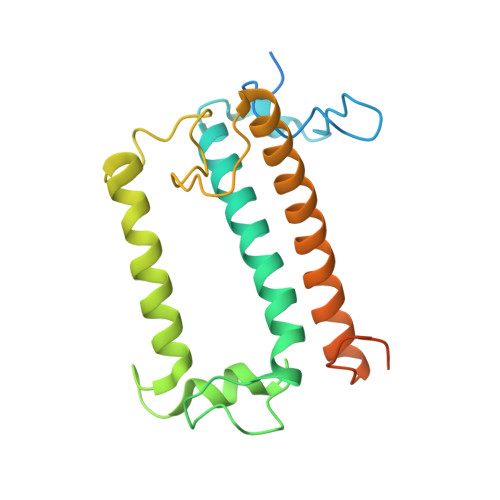
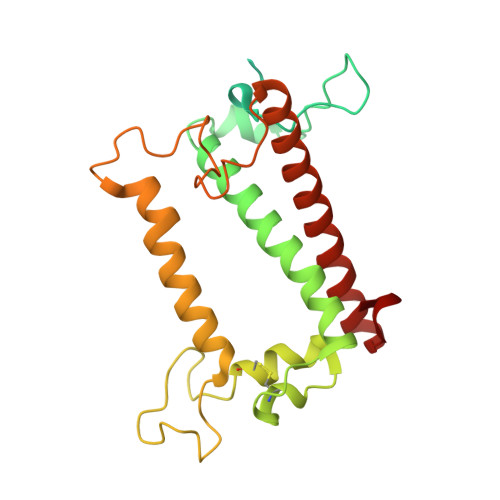
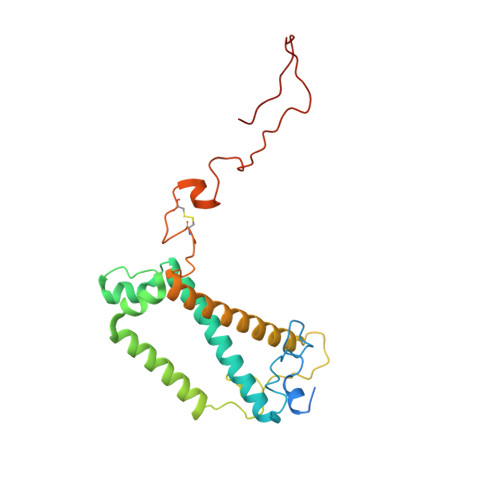
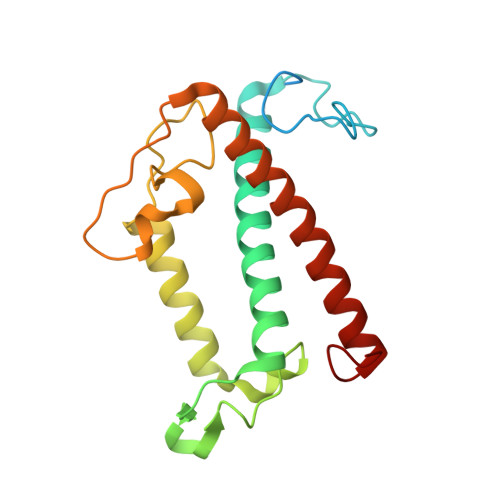
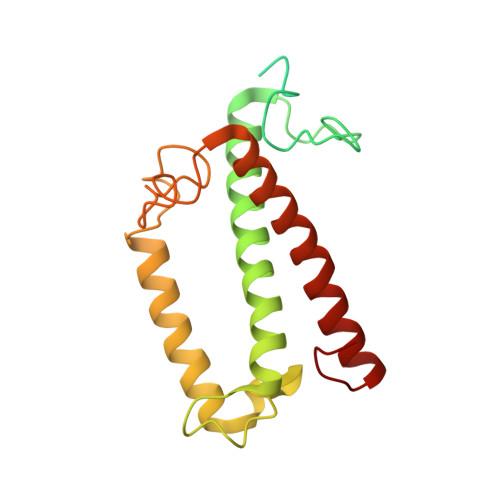
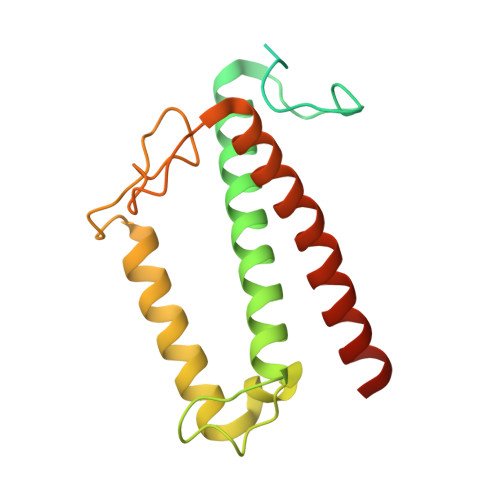
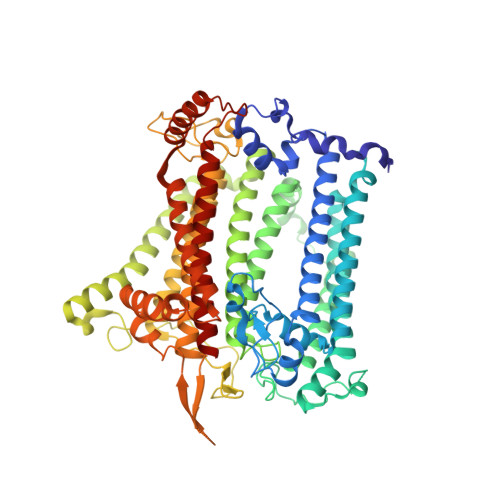
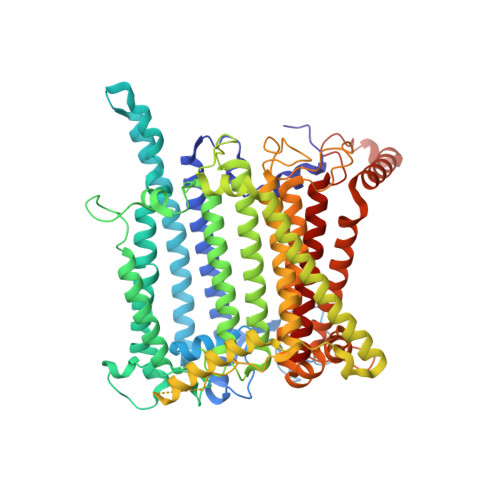
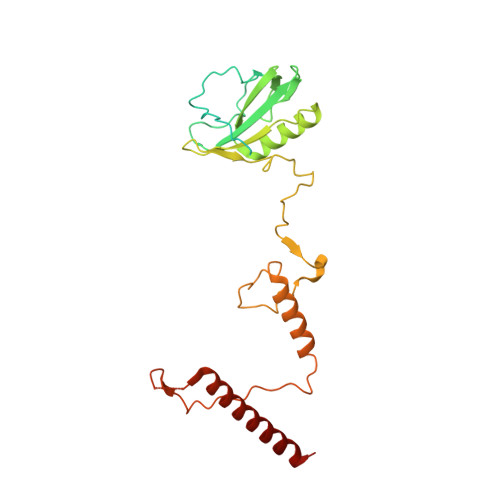
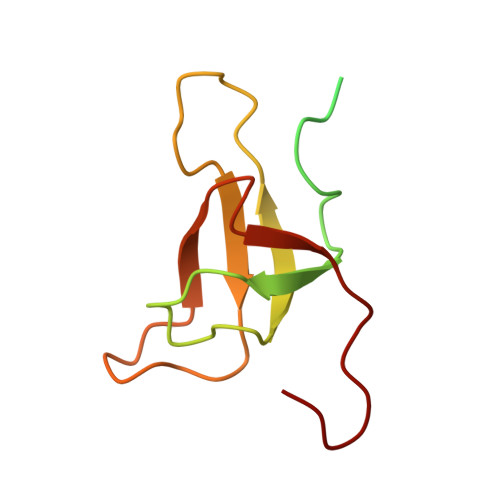
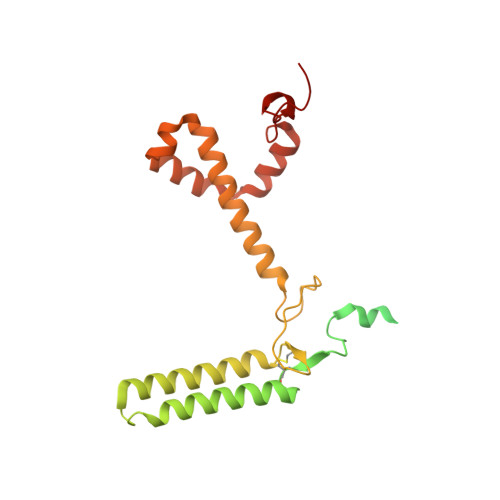
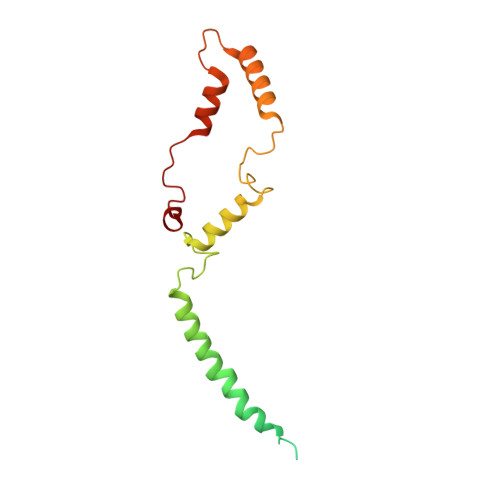
Architecture of symbiotic dinoflagellate photosystem I-light-harvesting supercomplex in Symbiodinium.
Zhao, L.S., Wang, N., Li, K., Li, C.Y., Guo, J.P., He, F.Y., Liu, G.M., Chen, X.L., Gao, J., Liu, L.N., Zhang, Y.Z.(2024) Nat Commun 15: 2392-2392
- PubMed: 38493166
- DOI: https://doi.org/10.1038/s41467-024-46791-x
- Primary Citation of Related Structures:
8JJR - PubMed Abstract:
Symbiodinium are the photosynthetic endosymbionts for corals and play a vital role in supplying their coral hosts with photosynthetic products, forming the nutritional foundation for high-yield coral reef ecosystems. Here, we determine the cryo-electron microscopy structure of Symbiodinium photosystem I (PSI) supercomplex with a PSI core composed of 13 subunits including 2 previously unidentified subunits, PsaT and PsaU, as well as 13 peridinin-Chl a/c-binding light-harvesting antenna proteins (AcpPCIs). The PSI-AcpPCI supercomplex exhibits distinctive structural features compared to their red lineage counterparts, including extended termini of PsaD/E/I/J/L/M/R and AcpPCI-1/3/5/7/8/11 subunits, conformational changes in the surface loops of PsaA and PsaB subunits, facilitating the association between the PSI core and peripheral antennae. Structural analysis and computational calculation of excitation energy transfer rates unravel specific pigment networks in Symbiodinium PSI-AcpPCI for efficient excitation energy transfer. Overall, this study provides a structural basis for deciphering the mechanisms governing light harvesting and energy transfer in Symbiodinium PSI-AcpPCI supercomplexes adapted to their symbiotic ecosystem, as well as insights into the evolutionary diversity of PSI-LHCI among various photosynthetic organisms.
- MOE Key Laboratory of Evolution and Marine Biodiversity, Frontiers Science Center for Deep Ocean Multispheres and Earth System & College of Marine Life Sciences, Ocean University of China, Qingdao, 266003, China.
Organizational Affiliation: