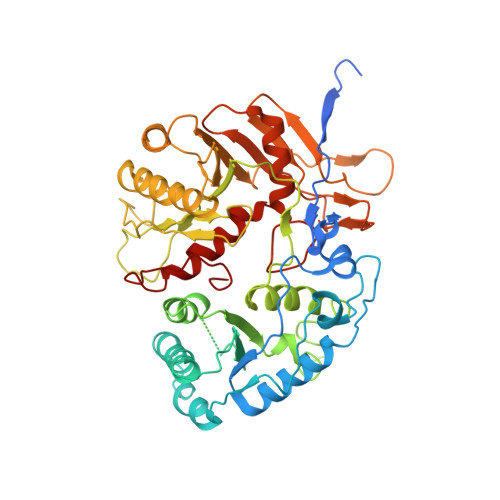
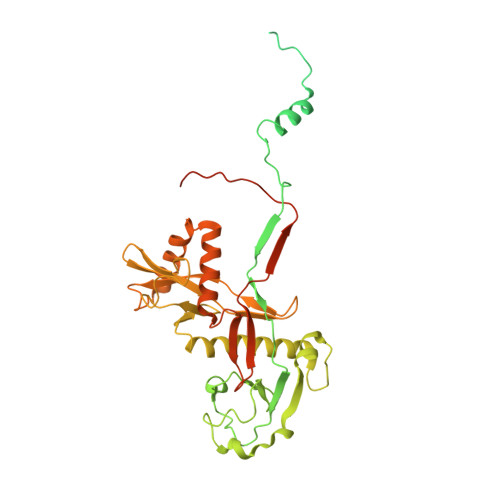
Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Guo, L., Huang, P., Li, Z., Shin, Y.C., Yan, P., Lu, M., Chen, M., Xiao, Y.(2024) Nat Chem Biol 20: 512-520
- PubMed: 37932527
- DOI: https://doi.org/10.1038/s41589-023-01478-0
- Primary Citation of Related Structures:
8J84, 8J8H, 8J9G, 8J9P, 8JAY - PubMed Abstract:
Short prokaryotic Ago accounts for most prokaryotic Argonaute proteins (pAgos) and is involved in defending bacteria against invading nucleic acids. Short pAgo associated with TIR-APAZ (SPARTA) has been shown to oligomerize and deplete NAD + upon guide-mediated target DNA recognition. However, the molecular basis of SPARTA inhibition and activation remains unknown. In this study, we determined the cryogenic electron microscopy structures of Crenotalea thermophila SPARTA in its inhibited, transient and activated states. The SPARTA monomer is auto-inhibited by its acidic tail, which occupies the guide-target binding channel. Guide-mediated target binding expels this acidic tail and triggers substantial conformational changes to expose the Ago-Ago dimerization interface. As a result, SPARTA assembles into an active tetramer, where the four TIR domains are rearranged and packed to form NADase active sites. Together with biochemical evidence, our results provide a panoramic vision explaining SPARTA auto-inhibition and activation and expand understanding of pAgo-mediated bacterial defense systems.
- State Key Laboratory of Natural Medicines, China Pharmaceutical University, Nanjing, China.
Organizational Affiliation: