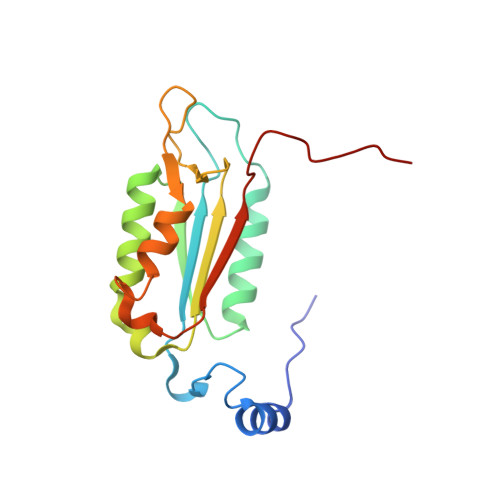
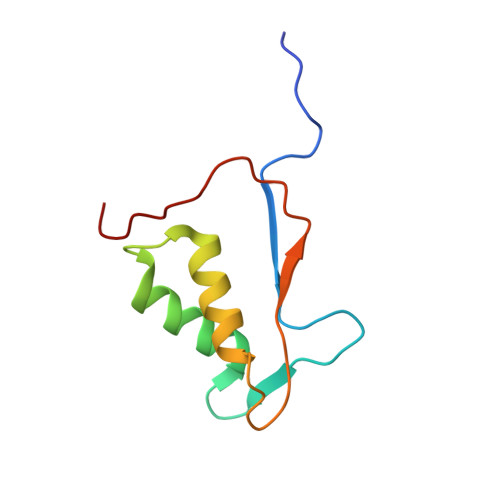
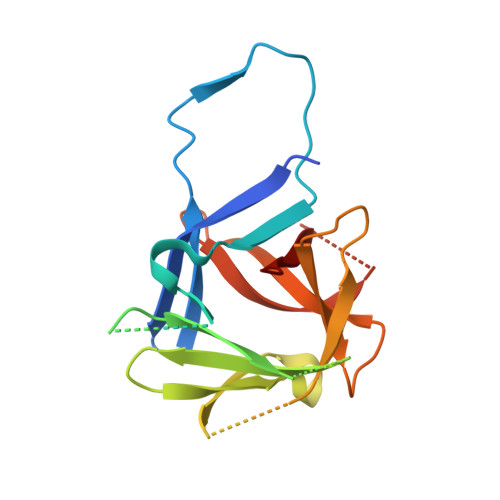
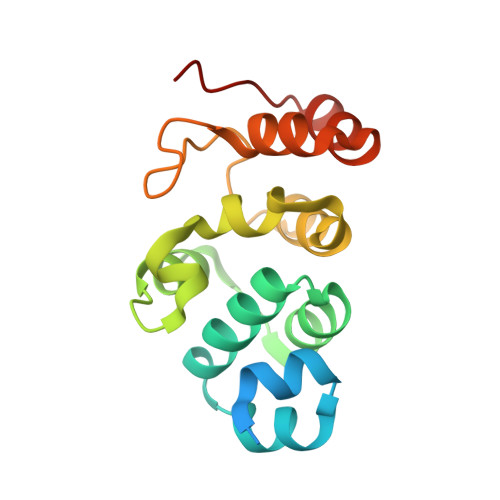
Recognition and maturation of IL-18 by caspase-4 noncanonical inflammasome.
Shi, X., Sun, Q., Hou, Y., Zeng, H., Cao, Y., Dong, M., Ding, J., Shao, F.(2023) Nature 624: 442-450
- PubMed: 37993714
- DOI: https://doi.org/10.1038/s41586-023-06742-w
- Primary Citation of Related Structures:
8J6K - PubMed Abstract:
The canonical (caspase-1) and noncanonical (comprising caspases 4, 5 and 11; hereafter, caspase-4/5/11) inflammasomes both cleave gasdermin D (GSDMD) to induce pyroptosis 1,2 . Whereas caspase-1 processes IL-1β and IL-18 for maturation 3-6 , no cytokine target has been firmly established for lipopolysaccharide-activated caspase-4/5/11 7-9 . Here we show that activated human caspase-4, but not mouse caspase-11, directly and efficiently processes IL-18 in vitro and during bacterial infections. Caspase-4 cleaves the same tetrapeptide site in pro-IL-18 as caspase-1. The crystal structure of the caspase-4-pro-IL-18 complex reveals a two-site (binary) substrate-recognition mechanism; the catalytic pocket engages the tetrapeptide, and a unique exosite that critically recognizes GSDMD 10 similarly binds to a specific structure formed jointly by the propeptide and post-cleavage-site sequences in pro-IL-18. This binary recognition is also used by caspase-5 as well as caspase-1 to process pro-IL-18. In caspase-11, a structural deviation around the exosite underlies its inability to target pro-IL-18, which is restored by rationally designed mutations. The structure of pro-IL-18 features autoinhibitory interactions between the propeptide and the post-cleavage-site region, preventing recognition by the IL-18Rα receptor. Cleavage by caspase-1, -4 or -5 induces substantial conformational changes of IL-18 to generate two critical receptor-binding sites. Our study establishes IL-18 as a target of lipopolysaccharide-activated caspase-4/5. The finding is paradigm shifting in the understanding of noncanonical-inflammasome-mediated defences and also the function of IL-18 in immunity and disease.
- National Institute of Biological Sciences, Beijing, Beijing, P. R. China.
Organizational Affiliation: