Functional characterization, structural basis, and regio-selectivity control of a promiscuous flavonoid 7,4'-di- O -glycosyltransferase from Ziziphus jujuba var. spinosa.
Wang, Z.L., Wei, W., Wang, H.D., Zhou, J.J., Wang, H.T., Chen, K., Wang, R.S., Li, F.D., Qiao, X., Zhou, H., Liang, Y., Ye, M.(2023) Chem Sci 14: 4418-4425
- PubMed: 37123177
- DOI: https://doi.org/10.1039/d2sc06504e
- Primary Citation of Related Structures:
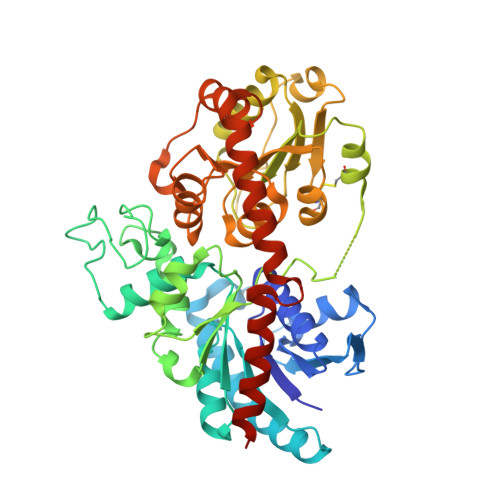
8INH - PubMed Abstract:
A highly efficient and promiscuous 7,4'-di- O -glycosyltransferase ZjOGT3 was discovered from the medicinal plant Ziziphus jujuba var. spinosa . ZjOGT3 could sequentially catalyse 4'- and 7- O -glycosylation of flavones to produce 7,4'-di- O -glycosides with obvious regio-selectivity. For 7,4'-dihydroxyl flavanones and 3- O -glycosylated 7,4'-dihydroxyl flavones, ZjOGT3 selectively catalyses 7- O -glycosylation. The crystal structure of ZjOGT3 was solved. Structural analysis, DFT calculations, MD simulations, and site-directed mutagenesis reveal that the regio-selectivity is mainly controlled by the enzyme microenvironment for 7,4'-dihydroxyl flavones and 3- O -glycosylated 7,4'-dihydroxyl flavones. For 7,4'-dihydroxyl flavanones, the selectivity is mainly controlled by intrinsic reactivity. ZjOGT3 is the first plant flavonoid 7,4'-di- O -glycosyltransferase with a crystal structure. This work could help understand the catalytic mechanisms of multi-site glycosyltransferases and provides an efficient approach to synthesise O -glycosides with medicinal potential.
- State Key Laboratory of Natural and Biomimetic Drugs, School of Pharmaceutical Sciences, Peking University 38 Xueyuan Road Beijing 100191 China yemin@bjmu.edu.cn.
Organizational Affiliation: