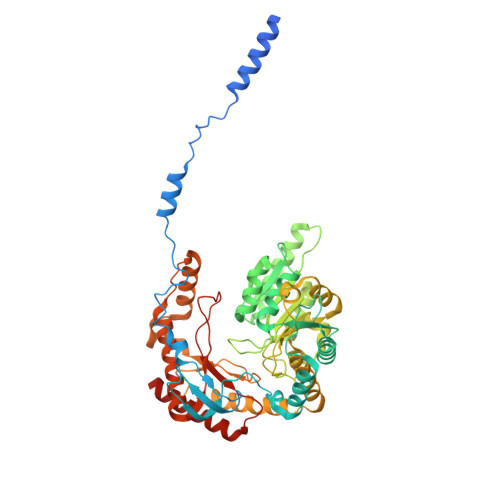
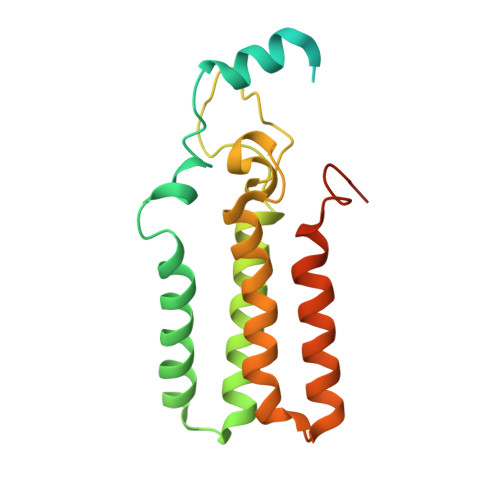
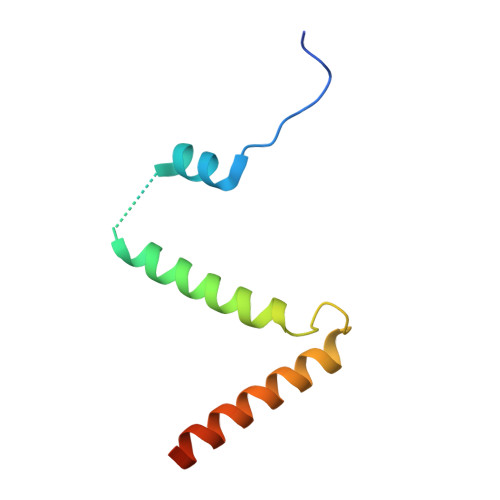
Collaborative regulation of yeast SPT-Orm2 complex by phosphorylation and ceramide.
Xie, T., Dong, F., Han, G., Wu, X., Liu, P., Zhang, Z., Zhong, J., Niranjanakumari, S., Gable, K., Gupta, S.D., Liu, W., Harrison, P.J., Campopiano, D.J., Dunn, T.M., Gong, X.(2024) Cell Rep 43: 113717-113717
- PubMed: 38285738
- DOI: https://doi.org/10.1016/j.celrep.2024.113717
- Primary Citation of Related Structures:
8IAJ, 8IAK, 8IAM - PubMed Abstract:
The homeostatic regulation of serine palmitoyltransferase (SPT) activity in yeast involves N-terminal phosphorylation of Orm proteins, while higher eukaryotes lack these phosphorylation sites. Although recent studies have indicated a conserved ceramide-mediated feedback inhibition of the SPT-ORM/ORMDL complex in higher eukaryotes, its conservation and relationship with phosphorylation regulation in yeast remain unclear. Here, we determine the structure of the yeast SPT-Orm2 complex in a dephosphomimetic state and identify an evolutionarily conserved ceramide-sensing site. Ceramide stabilizes the dephosphomimetic Orm2 in an inhibitory conformation, facilitated by an intramolecular β-sheet between the N- and C-terminal segments of Orm2. Moreover, we find that a phosphomimetic mutant of Orm2, positioned adjacent to its intramolecular β-sheet, destabilizes the inhibitory conformation of Orm2. Taken together, our findings suggest that both Orm dephosphorylation and ceramide binding are crucial for suppressing SPT activity in yeast. This highlights a distinctive regulatory mechanism in yeast involving the collaborative actions of phosphorylation and ceramide.
- Department of Chemical Biology, School of Life Sciences, Southern University of Science and Technology, Shenzhen, Guangdong 518055, China.
Organizational Affiliation: