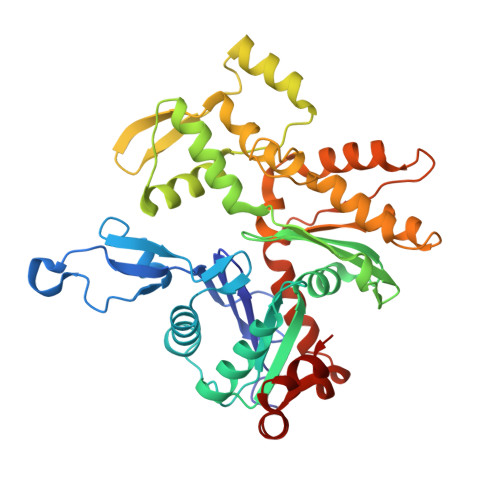
Structural basis of membrane skeleton organization in red blood cells.
Li, N., Chen, S., Xu, K., He, M.T., Dong, M.Q., Zhang, Q.C., Gao, N.(2023) Cell 186: 1912-1929.e18
- PubMed: 37044097
- DOI: https://doi.org/10.1016/j.cell.2023.03.017
- Primary Citation of Related Structures:
8IAH, 8IAI, 8IB2 - PubMed Abstract:
The spectrin-based membrane skeleton is a ubiquitous membrane-associated two-dimensional cytoskeleton underneath the lipid membrane of metazoan cells. Mutations of skeleton proteins impair the mechanical strength and functions of the membrane, leading to several different types of human diseases. Here, we report the cryo-EM structures of the native spectrin-actin junctional complex (from porcine erythrocytes), which is a specialized short F-actin acting as the central organizational unit of the membrane skeleton. While an α-/β-adducin hetero-tetramer binds to the barbed end of F-actin as a flexible cap, tropomodulin and SH3BGRL2 together create an absolute cap at the pointed end. The junctional complex is strengthened by ring-like structures of dematin in the middle actin layers and by patterned periodic interactions with tropomyosin over its entire length. This work serves as a structural framework for understanding the assembly and dynamics of membrane skeleton and offers insights into mechanisms of various ubiquitous F-actin-binding factors in other F-actin systems.
- State Key Laboratory of Membrane Biology, Peking-Tsinghua Joint Center for Life Sciences, School of Life Sciences, Peking University, Beijing 100871, China; Changping Laboratory, Beijing 102206, China. Electronic address: ningningli@pku.edu.cn.
Organizational Affiliation: