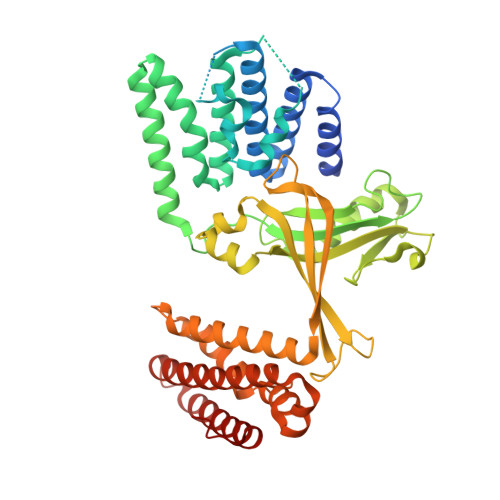
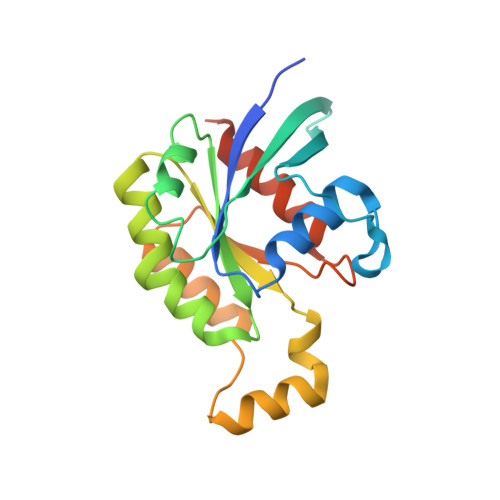
Structural basis for the dual GTPase specificity of the DOCK10 guanine nucleotide exchange factor.
Kukimoto-Niino, M., Ihara, K., Mishima-Tsumagari, C., Inoue, M., Fukui, Y., Yokoyama, S., Shirouzu, M.(2023) Biochem Biophys Res Commun 653: 12-20
- PubMed: 36848820
- DOI: https://doi.org/10.1016/j.bbrc.2023.02.054
- Primary Citation of Related Structures:
8I5F, 8I5V, 8I5W - PubMed Abstract:
Dedicator of cytokinesis 10 (DOCK10), an evolutionarily conserved guanine nucleotide exchange factor (GEF) for Rho GTPases, has the unique specificity within the DOCK-D subfamily to activate both Cdc42 and Rac, but the structural bases for these activities remained unknown. Here we present the crystal structures of the catalytic DHR2 domain of mouse DOCK10, complexed with either Cdc42 or Rac1. The structures revealed that DOCK10 DHR2 binds to Cdc42 or Rac1 by slightly changing the arrangement of its two catalytic lobes. DOCK10 also has a flexible binding pocket for the 56th GTPase residue, allowing a novel interaction with Trp56 Rac1 . The conserved residues in switch 1 of Cdc42 and Rac1 showed common interactions with the unique Lys-His sequence in the β5/β6 loop of DOCK10 DHR2 . However, the interaction of switch 1 in Rac1 was less stable than that of switch 1 in Cdc42, due to amino acid differences at positions 27 and 30. Structure-based mutagenesis identified the DOCK10 residues that determine the Cdc42/Rac1 dual specificity.
- Laboratory for Protein Functional and Structural Biology, RIKEN Center for Biosystems Dynamics Research, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa, 230-0045, Japan. Electronic address: kukimoto@riken.jp.
Organizational Affiliation: