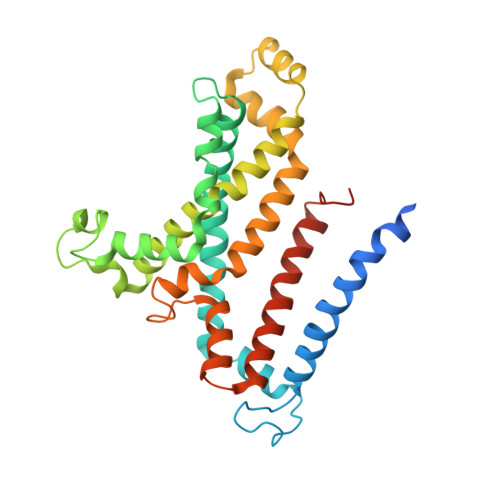
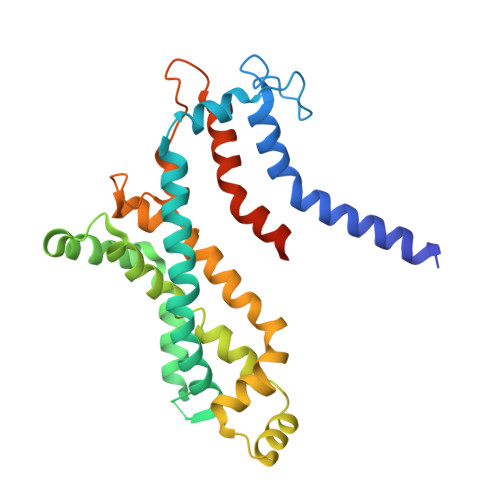
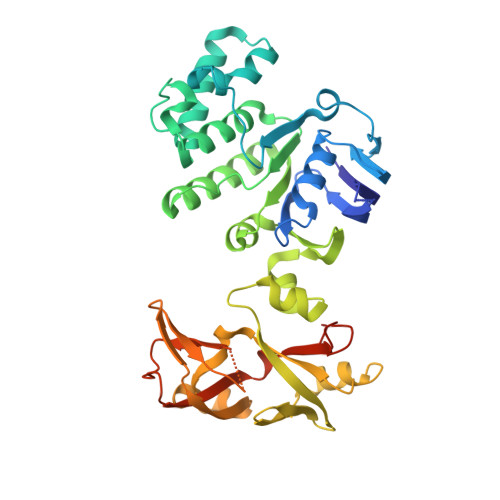
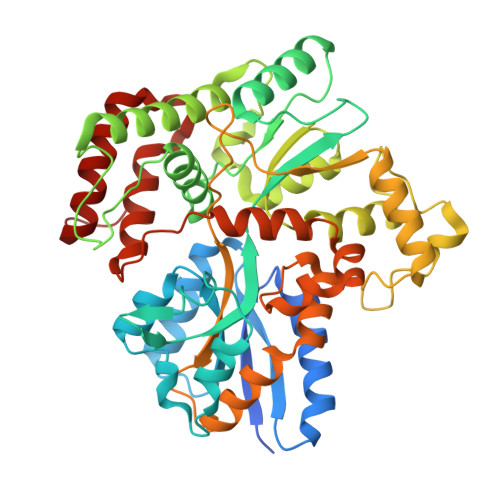
Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Liang, J., Yang, X., Hu, T., Gao, Y., Yang, Q., Yang, H., Peng, W., Zhou, X., Guddat, L.W., Zhang, B., Rao, Z., Liu, F.(2023) Structure 31: 1158-1165.e3
- PubMed: 37619560
- DOI: https://doi.org/10.1016/j.str.2023.07.014
- Primary Citation of Related Structures:
8HPL, 8HPM, 8HPN, 8HPR, 8HPS - PubMed Abstract:
The human pathogen, Mycobacterium tuberculosis (Mtb) relies heavily on trehalose for both survival and pathogenicity. The type I ATP-binding cassette (ABC) transporter LpqY-SugABC is the only trehalose import pathway in Mtb. Conformational dynamics of ABC transporters is an important feature to explain how they operate, but experimental structures are determined in a static environment. Therefore, a detailed transport mechanism cannot be elucidated because there is a lack of intermediate structures. Here, we used single-particle cryo-electron microscopy (cryo-EM) to determine the structure of the Mycobacterium smegmatis (M. smegmatis) trehalose-specific importer LpqY-SugABC complex in five different conformations. These structures have been classified and reconstructed from a single cryo-EM dataset. This study allows a comprehensive understanding of the trehalose recycling mechanism in Mycobacteria and also demonstrates the potential of single-particle cryo-EM to explore the dynamic structures of other ABC transporters and molecular machines.
- State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, China; Shanghai Institute for Advanced Immunochemical Studies and School of Life Science and Technology, ShanghaiTech University, Shanghai, China; Innovative Center for Pathogen Research, Guangzhou Laboratory, Guangzhou, China.
Organizational Affiliation: